An advanced bioinformatics approach for analyzing RNA-seq data reveals sigma H-dependent regulation of competence genes in Listeria monocytogenes
- PMID: 26880300
- PMCID: PMC4754846
- DOI: 10.1186/s12864-016-2432-9
An advanced bioinformatics approach for analyzing RNA-seq data reveals sigma H-dependent regulation of competence genes in Listeria monocytogenes
Abstract
Background: Alternative σ factors are important transcriptional regulators in bacteria. While σ(B) has been shown to control a large regulon and play important roles in stress response and virulence in the pathogen Listeria monocytogenes, the function of σ(H) has not yet been well defined in Listeria, even though σ(H) controls a large regulon in the closely related non-pathogenic Bacillus subtilis.
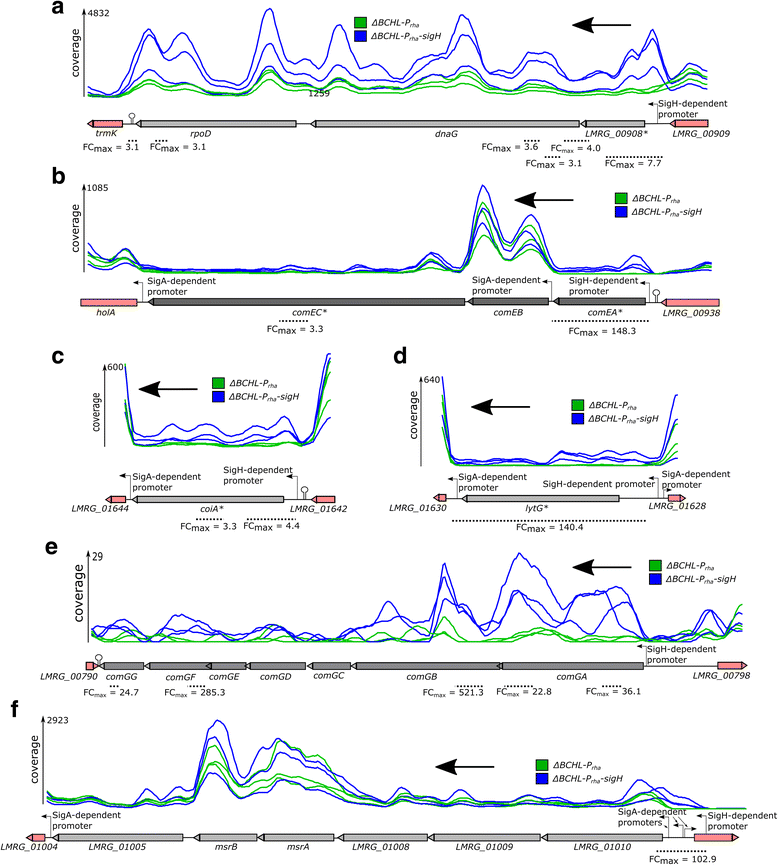
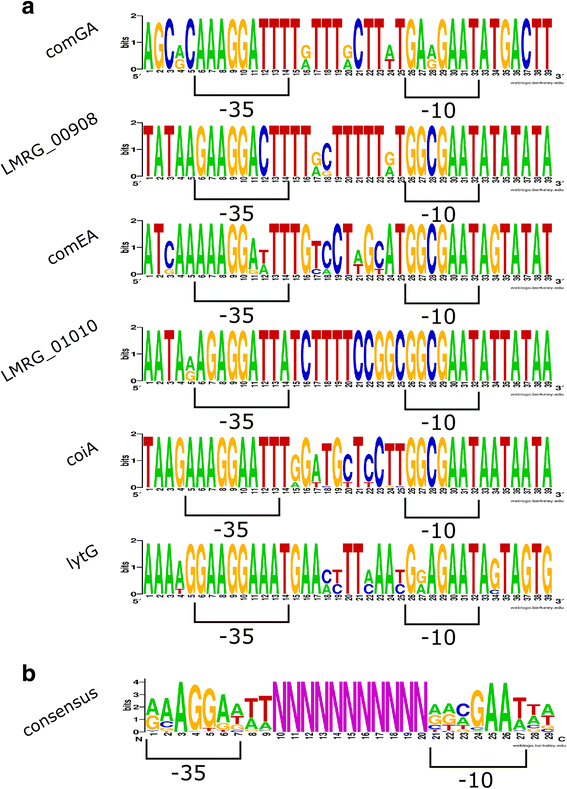
Results: Using RNA-seq characterization of a L. monocytogenes strain with deletions of all 4 genes encoding alternative σ factors (ΔBCHL), which was further modified to overexpress sigH (ΔBCHL::P rha -sigH), we identified 6 transcription units (TUs) that are transcribed from σ(H)-dependent promoters. Five of these TUs had not been previously identified. Identification of these promoters was facilitated by use of a bio-informatics approach that compared normalized RNA-seq coverage (NRC), between ΔBCHL::P rha -sigH and a ΔBCHL control, using sliding windows of 51 nt along the whole genome rather than comparing NRC calculated only for whole genes. Interestingly, we found that three operons that encode competence genes (comGABCDEFG, comEABC, coiA) are transcribed from σ(H)-dependent promoters. While these promoters were highly conserved in L. monocytogenes, none of them were found in all Listeria spp. and coiA and its σ(H)-dependent promoter were only found in L. monocytogenes.
Conclusions: Our data indicate that a number of L. monocytogenes competence genes are regulated by σ(H). This σ(H)-dependent regulation of competence related genes is conserved in the pathogen L. monocytogenes, but not in other non-pathogenic Listeria strains. Combined with prior data that indicated a role of σ(H) in virulence in a mouse model, this suggests a possible novel role of σ(H)-dependent competence genes in L. monocytogenes virulence. Development and implementation of a sliding window approach to identify differential transcription using RNA-seq data, not only allowed for identification of σ(H)-dependent promoters, but also provides a general approach for sensitive identification of differentially transcribed promoters and genes, particularly for genes that are transcribed from multiple promoter elements only some of which show differential transcription.
Figures

Similar articles
-
Home Alone: Elimination of All but One Alternative Sigma Factor in Listeria monocytogenes Allows Prediction of New Roles for σB.Front Microbiol. 2017 Oct 11;8:1910. doi: 10.3389/fmicb.2017.01910. eCollection 2017. Front Microbiol. 2017. PMID: 29075236 Free PMC article.
-
Listeria monocytogenes σH Contributes to Expression of Competence Genes and Intracellular Growth.J Bacteriol. 2016 Mar 31;198(8):1207-17. doi: 10.1128/JB.00718-15. Print 2016 Apr. J Bacteriol. 2016. PMID: 26833412 Free PMC article.
-
Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs.BMC Genomics. 2009 Dec 30;10:641. doi: 10.1186/1471-2164-10-641. BMC Genomics. 2009. PMID: 20042087 Free PMC article.
-
Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism.Future Microbiol. 2019 Jun;14:801-828. doi: 10.2217/fmb-2019-0072. Epub 2019 Jul 4. Future Microbiol. 2019. PMID: 31271064
-
[Contribution of sigma B to environmental stress tolerance in Listeria monocytogenes--a review].Wei Sheng Wu Xue Bao. 2009 Oct;49(10):1282-8. Wei Sheng Wu Xue Bao. 2009. PMID: 20069872 Review. Chinese.
Cited by
-
Roles of Alternative Sigma Factors in Invasion and Growth Characteristics of Listeria monocytogenes 10403S Into Human Epithelial Colorectal Adenocarcinoma Caco-2 Cell.Front Microbiol. 2022 Jul 14;13:901484. doi: 10.3389/fmicb.2022.901484. eCollection 2022. Front Microbiol. 2022. PMID: 35910626 Free PMC article.
-
Genome-wide identification of the context-dependent sRNA expression in Mycobacterium tuberculosis.BMC Genomics. 2020 Feb 18;21(1):167. doi: 10.1186/s12864-020-6573-5. BMC Genomics. 2020. PMID: 32070281 Free PMC article.
-
Natural DNA Transformation Is Functional in Lactococcus lactis subsp. cremoris KW2.Appl Environ Microbiol. 2017 Aug 1;83(16):e01074-17. doi: 10.1128/AEM.01074-17. Print 2017 Aug 15. Appl Environ Microbiol. 2017. PMID: 28625996 Free PMC article.
-
The Listeria monocytogenes Bile Stimulon under Acidic Conditions Is Characterized by Strain-Specific Patterns and the Upregulation of Motility, Cell Wall Modification Functions, and the PrfA Regulon.Front Microbiol. 2018 Feb 6;9:120. doi: 10.3389/fmicb.2018.00120. eCollection 2018. Front Microbiol. 2018. PMID: 29467736 Free PMC article.
-
Use of omics methods for the advancement of food quality and food safety.Anim Front. 2018 Nov 9;8(4):33-41. doi: 10.1093/af/vfy024. eCollection 2018 Oct. Anim Front. 2018. PMID: 32002228 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

