High-Resolution Mapping of the Folding Transition State of a WW Domain
- PMID: 26880334
- PMCID: PMC4835268
- DOI: 10.1016/j.jmb.2016.02.008
High-Resolution Mapping of the Folding Transition State of a WW Domain
Abstract
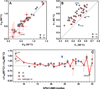
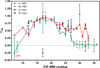
Fast-folding WW domains are among the best-characterized systems for comparing experiments and simulations of protein folding. Recent microsecond-resolution experiments and long duration (totaling milliseconds) single-trajectory modeling have shown that even mechanistic changes in folding kinetics due to mutation can now be analyzed. Thus, a comprehensive set of experimental data would be helpful to benchmark the predictions made by simulations. Here, we use T-jump relaxation in conjunction with protein engineering and report mutational Φ-values (Φ(M)) as indicators for folding transition-state structure of 65 side chain, 7 backbone hydrogen bond, and 6 deletion and /or insertion mutants within loop 1 of the 34-residue hPin1 WW domain. Forty-five cross-validated consensus mutants could be identified that provide structural constraints for transition-state structure within all substructures of the WW domain fold (hydrophobic core, loop 1, loop 2, β-sheet). We probe the robustness of the two hydrophobic clusters in the folding transition state, discuss how local backbone disorder in the native-state can lead to non-classical Φ(M)-values (Φ(M) > 1) in the rate-determining loop 1 substructure, and conclusively identify mutations and positions along the sequence that perturb the folding mechanism from loop 1-limited toward loop 2-limited folding.
Keywords: WW domain; folding transition state; laser T-jump; protein folding; Φ-value analysis.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures







Similar articles
-
Influence of hPin1 WW N-terminal domain boundaries on function, protein stability, and folding.Protein Sci. 2007 Jul;16(7):1495-501. doi: 10.1110/ps.072775507. Protein Sci. 2007. PMID: 17586778 Free PMC article.
-
Mapping the transition state of the WW domain beta-sheet.J Mol Biol. 2000 Apr 28;298(2):283-92. doi: 10.1006/jmbi.2000.3665. J Mol Biol. 2000. PMID: 10764597
-
Temperature-dependent folding pathways of Pin1 WW domain: an all-atom molecular dynamics simulation of a Gō model.Biophys J. 2007 Sep 15;93(6):2152-61. doi: 10.1529/biophysj.106.102095. Epub 2007 May 18. Biophys J. 2007. PMID: 17513360 Free PMC article.
-
Understanding the mechanism of beta-sheet folding from a chemical and biological perspective.Biopolymers. 2008;90(6):751-8. doi: 10.1002/bip.21101. Biopolymers. 2008. PMID: 18844292 Review.
-
Beta-sheet folding mechanisms from perturbation energetics.Curr Opin Struct Biol. 2006 Feb;16(1):94-101. doi: 10.1016/j.sbi.2006.01.014. Epub 2006 Jan 25. Curr Opin Struct Biol. 2006. PMID: 16442278 Review.
Cited by
-
Atomic-level thermodynamics analysis of the binding free energy of SARS-CoV-2 neutralizing antibodies.Proteins. 2023 May;91(5):694-704. doi: 10.1002/prot.26458. Epub 2023 Jan 1. Proteins. 2023. PMID: 36564921 Free PMC article.
-
New Insights into Folding, Misfolding, and Nonfolding Dynamics of a WW Domain.J Phys Chem B. 2020 May 14;124(19):3855-3872. doi: 10.1021/acs.jpcb.0c00628. Epub 2020 May 1. J Phys Chem B. 2020. PMID: 32271570 Free PMC article.
-
Using Cooperatively Folded Peptides To Measure Interaction Energies and Conformational Propensities.Acc Chem Res. 2017 Aug 15;50(8):1875-1882. doi: 10.1021/acs.accounts.7b00195. Epub 2017 Jul 19. Acc Chem Res. 2017. PMID: 28723063 Free PMC article.
-
The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.J Am Chem Soc. 2016 Jun 22;138(24):7636-48. doi: 10.1021/jacs.6b02879. Epub 2016 Jun 14. J Am Chem Soc. 2016. PMID: 27249581 Free PMC article.
-
Identification of novel functional mini-receptors by combinatorial screening of split-WW domains.Chem Sci. 2022 Jul 14;13(31):9079-9090. doi: 10.1039/d2sc01078j. eCollection 2022 Aug 10. Chem Sci. 2022. PMID: 36091217 Free PMC article.
References
-
- Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr. Opin. Struct. Biol. 2004;14:76–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

