High-Resolution Mapping of the Folding Transition State of a WW Domain
- PMID: 26880334
- PMCID: PMC4835268
- DOI: 10.1016/j.jmb.2016.02.008
High-Resolution Mapping of the Folding Transition State of a WW Domain
Abstract
Fast-folding WW domains are among the best-characterized systems for comparing experiments and simulations of protein folding. Recent microsecond-resolution experiments and long duration (totaling milliseconds) single-trajectory modeling have shown that even mechanistic changes in folding kinetics due to mutation can now be analyzed. Thus, a comprehensive set of experimental data would be helpful to benchmark the predictions made by simulations. Here, we use T-jump relaxation in conjunction with protein engineering and report mutational Φ-values (Φ(M)) as indicators for folding transition-state structure of 65 side chain, 7 backbone hydrogen bond, and 6 deletion and /or insertion mutants within loop 1 of the 34-residue hPin1 WW domain. Forty-five cross-validated consensus mutants could be identified that provide structural constraints for transition-state structure within all substructures of the WW domain fold (hydrophobic core, loop 1, loop 2, β-sheet). We probe the robustness of the two hydrophobic clusters in the folding transition state, discuss how local backbone disorder in the native-state can lead to non-classical Φ(M)-values (Φ(M) > 1) in the rate-determining loop 1 substructure, and conclusively identify mutations and positions along the sequence that perturb the folding mechanism from loop 1-limited toward loop 2-limited folding.
Keywords: WW domain; folding transition state; laser T-jump; protein folding; Φ-value analysis.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures





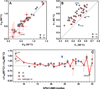


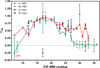
References
-
- Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr. Opin. Struct. Biol. 2004;14:76–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

