Functional screen identifies regulators of murine hematopoietic stem cell repopulation
- PMID: 26880577
- PMCID: PMC4813668
- DOI: 10.1084/jem.20150806
Functional screen identifies regulators of murine hematopoietic stem cell repopulation
Erratum in
-
Correction: Functional screen identifies regulators of murine hematopoietic stem cell repopulation.J Exp Med. 2016 Oct 17;213(11):2525. doi: 10.1084/jem.2015080609232016c. Epub 2016 Sep 27. J Exp Med. 2016. PMID: 27679570 Free PMC article. No abstract available.
Abstract
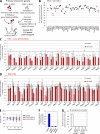
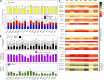
Understanding the molecular regulation of hematopoietic stem and progenitor cell (HSPC) engraftment is paramount to improving transplant outcomes. To discover novel regulators of HSPC repopulation, we transplanted >1,300 mice with shRNA-transduced HSPCs within 24 h of isolation and transduction to focus on detecting genes regulating repopulation. We identified 17 regulators of HSPC repopulation: Arhgef5, Armcx1, Cadps2, Crispld1, Emcn, Foxa3, Fstl1, Glis2, Gprasp2, Gpr56, Myct1, Nbea, P2ry14, Smarca2, Sox4, Stat4, and Zfp251. Knockdown of each of these genes yielded a loss of function, except in the cases of Armcx1 and Gprasp2, whose loss enhanced hematopoietic stem cell (HSC) repopulation. The discovery of multiple genes regulating vesicular trafficking, cell surface receptor turnover, and secretion of extracellular matrix components suggests active cross talk between HSCs and the niche and that HSCs may actively condition the niche to promote engraftment. We validated that Foxa3 is required for HSC repopulating activity, as Foxa3(-/-) HSC fails to repopulate ablated hosts efficiently, implicating for the first time Foxa genes as regulators of HSPCs. We further show that Foxa3 likely regulates the HSC response to hematologic stress. Each gene discovered here offers a window into the novel processes that regulate stable HSPC engraftment into an ablated host.
© 2016 Holmfeldt et al.
Figures





References
-
- Boitano A.E., Wang J., Romeo R., Bouchez L.C., Parker A.E., Sutton S.E., Walker J.R., Flaveny C.A., Perdew G.H., Denison M.S., et al. . 2010. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 329:1345–1348. 10.1126/science.1191536 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

