Validation of Housekeeping Genes to Study Human Gingival Stem Cells and Their In Vitro Osteogenic Differentiation Using Real-Time RT-qPCR
- PMID: 26880978
- PMCID: PMC4736224
- DOI: 10.1155/2016/6261490
Validation of Housekeeping Genes to Study Human Gingival Stem Cells and Their In Vitro Osteogenic Differentiation Using Real-Time RT-qPCR
Abstract
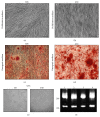
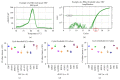
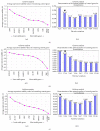
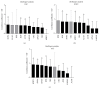
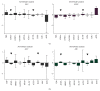
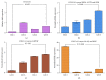
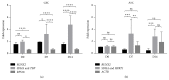
Gingival stem cells (GSCs) are recently isolated multipotent cells. Their osteogenic capacity has been validated in vitro and may be transferred to human cell therapy for maxillary large bone defects, as they share a neural crest cell origin with jaw bone cells. RT-qPCR is a widely used technique to study gene expression and may help us to follow osteoblast differentiation of GSCs. For accurate results, the choice of reliable housekeeping genes (HKGs) is crucial. The aim of this study was to select the most reliable HKGs for GSCs study and their osteogenic differentiation (dGSCs). The analysis was performed with ten selected HKGs using four algorithms: ΔCt comparative method, GeNorm, BestKeeper, and NormFinder. This study demonstrated that three HKGs, SDHA, ACTB, and B2M, were the most stable to study GSC, whereas TBP, SDHA, and ALAS1 were the most reliable to study dGSCs. The comparison to stem cells of mesenchymal origin (ASCs) showed that SDHA/HPRT1 were the most appropriate for ASCs study. The choice of suitable HKGs for GSCs is important as it gave access to an accurate analysis of osteogenic differentiation. It will allow further study of this interesting stem cells source for future human therapy.
Figures

Similar articles
-
RPLP0/TBP are the most stable reference genes for human dental pulp stem cells under osteogenic differentiation.World J Stem Cells. 2024 Jun 26;16(6):656-669. doi: 10.4252/wjsc.v16.i6.656. World J Stem Cells. 2024. PMID: 38948092 Free PMC article.
-
Exploring transcriptomic databases to identify and experimentally validate tissue-specific consensus reference gene for gene expression normalization in BALB/c mice acutely exposed to 2,3,7,8-Tetrachlorodibenzo-p-dioxin.Curr Res Toxicol. 2025 Apr 27;8:100234. doi: 10.1016/j.crtox.2025.100234. eCollection 2025. Curr Res Toxicol. 2025. PMID: 40391131 Free PMC article.
-
Housekeeping Gene Stability in Adipose Mesenchymal Stromal Cells Cultivated in Serum/Xeno-Free Media for Osteoarthritis.Cells. 2024 Jan 16;13(2):167. doi: 10.3390/cells13020167. Cells. 2024. PMID: 38247858 Free PMC article.
-
Identification of Appropriate Housekeeping Genes for Gene Expression Analysis in Long-term Hypoxia-treated Kidney Cells.Adv Biomed Res. 2017 Feb 22;6:15. doi: 10.4103/2277-9175.200790. eCollection 2017. Adv Biomed Res. 2017. PMID: 28299307 Free PMC article.
-
Evaluation of housekeeping genes for quantitative gene expression analysis in the equine kidney.J Equine Sci. 2016;27(4):165-168. doi: 10.1294/jes.27.165. Epub 2016 Dec 15. J Equine Sci. 2016. PMID: 27974876 Free PMC article.
Cited by
-
Selective Blockade of Two Aquaporin Channels, AQP3 and AQP9, Impairs Human Leukocyte Migration.Cells. 2025 Jun 11;14(12):880. doi: 10.3390/cells14120880. Cells. 2025. PMID: 40558507 Free PMC article.
-
Head to Knee: Cranial Neural Crest-Derived Cells as Promising Candidates for Human Cartilage Repair.Stem Cells Int. 2019 Jan 15;2019:9310318. doi: 10.1155/2019/9310318. eCollection 2019. Stem Cells Int. 2019. PMID: 30766608 Free PMC article. Review.
-
RPLP0/TBP are the most stable reference genes for human dental pulp stem cells under osteogenic differentiation.World J Stem Cells. 2024 Jun 26;16(6):656-669. doi: 10.4252/wjsc.v16.i6.656. World J Stem Cells. 2024. PMID: 38948092 Free PMC article.
-
Gene Expression Profile and Acute Gene Expression Response to Sclerostin Inhibition in Osteogenesis Imperfecta Bone.JBMR Plus. 2020 Jul 4;4(8):e10377. doi: 10.1002/jbm4.10377. eCollection 2020 Aug. JBMR Plus. 2020. PMID: 32803109 Free PMC article.
-
Comparative study of the osteogenic potential of mesenchymal stem cells derived from different sources.J Clin Exp Dent. 2018 Jan 1;10(1):e7-e13. doi: 10.4317/jced.53957. eCollection 2018 Jan. J Clin Exp Dent. 2018. PMID: 29670709 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

