GemC1 controls multiciliogenesis in the airway epithelium
- PMID: 26882546
- PMCID: PMC4772991
- DOI: 10.15252/embr.201540882
GemC1 controls multiciliogenesis in the airway epithelium
Abstract
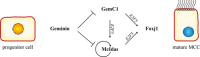
Multiciliated cells are terminally differentiated, post-mitotic cells that form hundreds of motile cilia on their apical surface. Defects in multiciliated cells lead to disease, including mucociliary clearance disorders that result from ciliated cell disfunction in airways. The pathway controlling multiciliogenesis, however, remains poorly characterized. We showed that GemC1, previously implicated in cell cycle control, is a central regulator of ciliogenesis. GemC1 is specifically expressed in ciliated epithelia. Ectopic expression of GemC1 is sufficient to induce early steps of multiciliogenesis in airway epithelial cells ex vivo, upregulating McIdas and FoxJ1, key transcriptional regulators of multiciliogenesis. GemC1 directly transactivates the McIdas and FoxJ1 upstream regulatory sequences, and its activity is enhanced by E2F5 and inhibited by Geminin. GemC1-knockout mice are born with airway epithelia devoid of multiciliated cells. Our results identify GemC1 as an essential regulator of ciliogenesis in the airway epithelium and a candidate gene for mucociliary disorders.
Keywords: McIdas; cell cycle; ciliary epithelia; ciliopathies; respiratory disorders.
© 2016 The Authors.
Figures
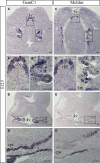
In situ hybridization for GemC1 and McIdas mRNA was performed on sagittal sections of E10.5 dpc mouse brain. GemC1 and McIdas mRNA is specifically detected in the developing choroid plexuses of the ventricles. Lower panels show higher magnifications. Scale bars, 100 μm.
In situ hybridization on sagittal (upper) and coronal (middle and lower) sections of the airways of E15.5 dpc mouse embryos, depicting specific GemC1 and McIdas mRNA expression in the respiratory epithelium of the nasal and oral cavity and in the upper bronchial tree in the mouse lungs. Inserts show higher‐magnification images. Scale bars, 200 μm.
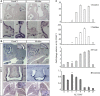
MTECs were cultured ex vivo and transitioned to ALI culture condition. Quantitative real‐time PCR was used to measure the expression levels of GemC1, McIdas, FoxJ1 and Geminin mRNA levels at different time points during multiciliogenesis. Values indicate expression levels (+/− standard error, s.e.) relative to ALI DAY 1 (marked by *), following normalization against the housekeeping HPRT mRNA. qPCR data were analyzed by the REST‐MCS beta software from at least three independent experiments.
- A–D
In situ hybridization of GemC1 (A‐B') and McIdas (C‐D') mRNA on coronal brain sections of E12.5 dpc mouse embryos. GemC1 and McIdas mRNAs are specifically detected in the developing choroid plexuses of the lateral and third ventricles (A–A″ and C–C″, respectively) and of the fourth ventricle (B, B′ and D, D′, respectively). In the fourth ventricle, McIdas expression is confined to the base of the choroid plexus. Abbreviations: lv: lateral ventricles, 3v: third ventricle, 4v: fourth ventricle, cpe: choroid plexus epithelium, ch: cortical hem, Eth: eminentia thalami. Scale bars, 100 μm.
- A–D
In situ hybridization, using probes for GemC1 (A–B) and McIdas (C–D) mRNA, was performed on coronal sections of E14.5 dpc mouse embryos. GemC1 expression is specifically detected throughout the choroid plexus epithelium in all the ventricles. McIdas expression is markedly reduced by this stage in all ventricles, with scattered positive cells at the base of the choroid plexus and eminentia thalami. Abbreviations: lv: lateral ventricles, 3v: third ventricle, Eth: eminentia thalami, 4v: fourth ventricle, cpe: choroid plexus epithelium. Scale bars, 100 μm.
- A, B
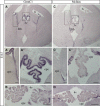
In situ hybridization for GemC1 (A–A″) and McIdas (B–B″) mRNA on sagittal sections of E18.5 dpc mouse lungs was performed, followed by immunofluorescence with an anti‐Foxj1‐specific antibody (green). GemC1 and McIdas mRNAs are specifically detected in the developing mouse distal airway and co‐expressed with Foxj1. Scale bars, 10 μm.
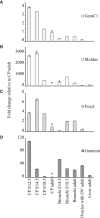
- A–D
GemC1 (A) quantitative mRNA expression analysis using real‐time PCR (qPCR) was performed in various tissues from mouse embryos and adult mice in parallel with McIdas (B), Foxj1 (C), and Geminin (D). Values indicate expression levels (+/− s.d.) relative to choroid plexus adult values set as 1 (*), following normalization against HPRT mRNA levels, which served as an internal control. qPCR data were analyzed by the REST‐MCS beta software from at least three independent experiments. Abbreviations: CP: choroid plexus, OV: oviducts.
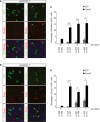
- A, B
MTEC cultures were infected with a lentivirus expressing either GFP‐GemC1 (GemC1) or GFP as a control and two days later transitioned to ALI culture conditions. Cells harvested at different time points during differentiation were co‐stained with antibodies against GFP (green) to mark infected cells and endogenous McIdas (A) or Foxj1 (B) (red). Arrows indicate infected cells that are McIdas or Foxj1 positive. Scale bars, 10 μm.
- C, D
The percentage of infected cells expressing McIdas (C) or Foxj1 (D) at each time point was quantified. Data are presented as the mean values of at least 10 independent fields obtained from at least two independent experiments for each condition. Error bars indicate ± SEM. *P < 0.05, ***P < 0.001. P‐values were calculated by the nonparametric two‐tailed Mann–Whitney test.
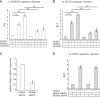
- A–D
MCIDAS (A) and FOXJ1 (B, D) regulatory elements (see Materials and Methods) cloned upstream of the luciferase gene were co‐transfected into HEK293T cells with vectors expressing GemC1, McIdas and Geminin, as indicated or an empty vector (−) as a control. In (D), McIdas siRNAs or a control siRNA were also co‐transfected. McIdas mRNA levels after McIdas RNAi were assessed by qPCR (C). All luciferase experiments (A, B, D) were normalized for transfection efficiency with an expression vector for Renilla luciferase. Fold induction is the ratio between the normalized luciferase activity induced by the expression constructs and that induced by the empty expression vector. Data are the mean values of at least three independent experiments, and error bars indicate ± SEM. **P < 0.01. P‐values were calculated by the nonparametric two‐tailed Mann–Whitney test. Abbreviations: RLF: relative luciferase fold induction.
- A, B
E2F family members E2F1, E2F4, and E2F5 examined for their transactivation activity on MCIDAS (A) and FOXJ1 (B) promoters. Vectors expressing each one of the three E2F members, or an empty vector (−) as a control, were co‐transfected into HEK293T cells with either the MCIDAS (A) or the FOXJ1 (B) regulatory elements cloned upstream from a luciferase reporter. While E2F5 shows no transactivation activity on its own, E2F4 exhibits high transactivation activity on both promoters.
- C, D
An E2F1‐expressing vector was co‐transfected with vectors expressing GemC1, McIdas, Geminin, or an empty vector (−) as a control and the MCIDAS (C) or FOXJ1 (D) regulatory elements cloned upstream of a luciferase reporter into HEK293T cells. The transactivation activity of E2F1 on these promoters is unaffected by GemC1, McIdas, or Geminin co‐expression.
- E
GEMC1 upstream regulatory elements were co‐transfected with vectors expressing McIdas, E2F5 or an empty vector (−) as a control into HEK293T cells. McIdas cannot transactivate the GEMC1 regulatory elements, either alone or with E2F5. Transactivation of MCIDAS regulatory elements is plotted in parallel for comparison.
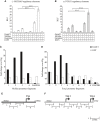
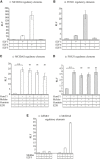
- A, B
MCIDAS (A) and FOXJ1 (B) regulatory elements cloned upstream of the luciferase gene were co‐transfected into HEK293T cells with vectors expressing GemC1, McIdas, Geminin and E2F5, as indicated, or an empty vector (−) as a control. Luciferase values were normalized against co‐transfected Renilla luciferase and depicted as fold induction over the control empty vector. Data shown are mean values of at least three independent experiments, and error bars indicate ± SEM. ***P < 0.001. P‐values were calculated by the nonparametric two‐tailed Mann–Whitney test. Abbreviations: RLF: relative luciferase fold induction.
- C, D
Chromatin immunoprecipitation of MCIDAS and FOXJ1 promoter fragments by GemC1. HEK293T cells were co‐transfected with vectors expressing GemC1‐GFP (GemC1) or GFP as a control, E2F5 and DP1, together with the regulatory elements of either MCIDAS (C) or FOXJ1 (D). Following chromatin immunoprecipitation, DNA fragments of the MCIDAS and FOXJ1 promoters were detected in immunoprecipitates by qPCR. Data from a representative experiment are shown as fold enrichment of MCIDAS and FOXJ1 promoter fragments in anti‐GFP immunoprecipitates over control IgG immunoprecipitates. At least two independent experiments were performed.
- E, F
Schematic representation of the promoter fragments from the MCIDAS (E) and FOXJ1 (F) regulatory elements assayed by chromatin immunoprecipitation. For FOXJ1, two putative transcription start sites (TSS) have been proposed and are indicated (see Materials and Methods).

Schematic representation of the wild‐type (WT) and knockout (KO) GemC1 alleles. A lacZ‐neo cassette has been inserted between exons 2 and 3 of the GemC1 gene. A splicing acceptor site (SA) upstream of lacZ ablates gene function (knockout‐first allele, 38)
GemC1KO/KO mice compared to their wild‐type littermates (GemC1KO/WT) at P5.
Survival plot of mice homozygous for the GemC1 deletion (GemC1KO/KO, n = 8) and control wild‐type or heterozygous littermates GemC1 (GemC1WT, n = 14). GemC1‐knockout mice exhibit markedly reduced growth after birth and die within the first postnatal week.
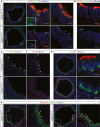
- A, B
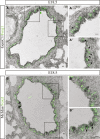
Transverse sections from trachea isolated from GemC1WT/WT (upper) or GemC1KO/KO (lower) P0 mice, immunostained with antibodies against acetylated α‐tubulin (ACT, cilia marker, red), pericentrin (PCNT, marker for nascent centrioles, green), γ‐tubulin (centriole marker, green) and E‐cadherin (marker for cell boundaries, blue). Serial images have been merged to show the complete trachea section (left). Inserts show higher‐magnification images for acetylated α‐tubulin and pericentrin staining in GemC1WT/WT and GemC1KO/KO trachea, respectively. Scale bars, 10 μm.
- C
McIdas‐ and Foxj1‐specific antibodies were used for immunofluorescence on trachea from GemC1WT/WT (upper) or GemC1KO/KO (lower) P0 mice. Arrows show representative positive cells for McIdas (green) and FoxJ1 (red) in the GemC1WT/WT mice. DNA was stained with Draq‐5 (blue). Scale bars, 10 μm.
- D
Transverse sections from P0 GemC1WT/WT (upper) or GemC1KO/KO (lower) trachea, immunostained with antibodies against keratin 5 (Krt5, basal cell marker, green) and acetylated α‐tubulin (ACT, cilia marker, red). DNA was stained with Draq‐5 (blue). Serial images have been merged to show the complete trachea section on the left. Higher‐magnification images are shown on the right. Scale bars, 10 μm.
- E
Transverse sections from P0 GemC1WT/WT (left two images) or GemC1KO/KO (right two images) trachea, immunostained with antibodies against Muc5ac (marker for mucus‐expressing cells, red) and E‐cadherin (marker for cell boundaries, green). DNA was stained with Draq‐5 (blue). Serial images have been merged to show the complete trachea section (left). Higher magnifications from the same section are shown on the right. Scale bars, 10 μm.

- A–C
Quantitative mRNA expression analysis, using real‐time PCR, was performed on E16.5 mouse tracheae isolated from either GemC1WT/WT or GemC1KO/KO mice. For each condition, tracheae from 3 embryos were pooled and analyzed in duplicate for the mRNA expression levels of McIdas (A), Foxj1 (B), and Geminin (C). Values indicate expression levels (+/− s.e.) relative to GemC1WT/WT values set as 1, following normalization against HPRT mRNA levels, which served as an internal control. qPCR data were analyzed by the REST‐MCS beta software.
Comment in
-
It's a family act: the geminin triplets take center stage in motile ciliogenesis.EMBO J. 2016 May 2;35(9):904-6. doi: 10.15252/embj.201694206. Epub 2016 Mar 22. EMBO J. 2016. PMID: 27006276 Free PMC article.
References
-
- Ishikawa H, Marshall WF (2011) Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol 12: 222–234 - PubMed
-
- Fliegauf M, Benzing T, Omran H (2007) When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8: 880–893 - PubMed
-
- Rock JR, Hogan BL (2011) Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol 27: 493–512 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

