Spatiotemporal hierarchy in antibody recognition against transmitted HIV-1 envelope glycoprotein during natural infection
- PMID: 26883323
- PMCID: PMC4756523
- DOI: 10.1186/s12977-016-0243-3
Spatiotemporal hierarchy in antibody recognition against transmitted HIV-1 envelope glycoprotein during natural infection
Abstract
Background: Majority of HIV-1 infection is established by one transmitted/founder virus and understanding how the neutralizing antibodies develop against this virus is critical for our rational design an HIV-1 vaccine.
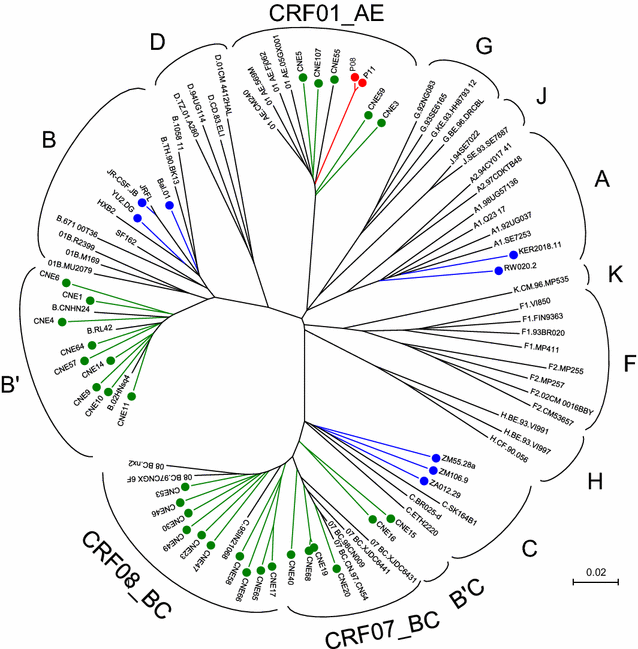
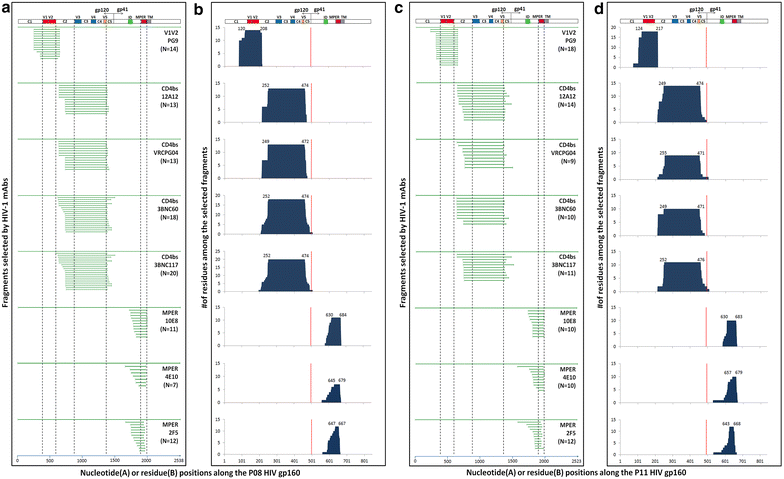
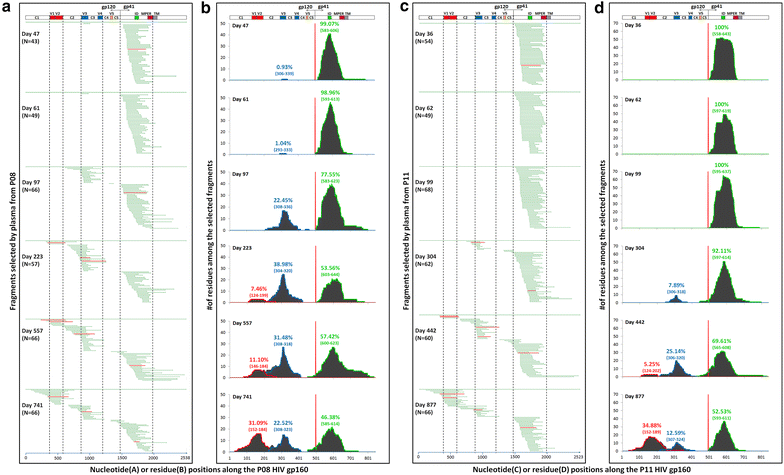
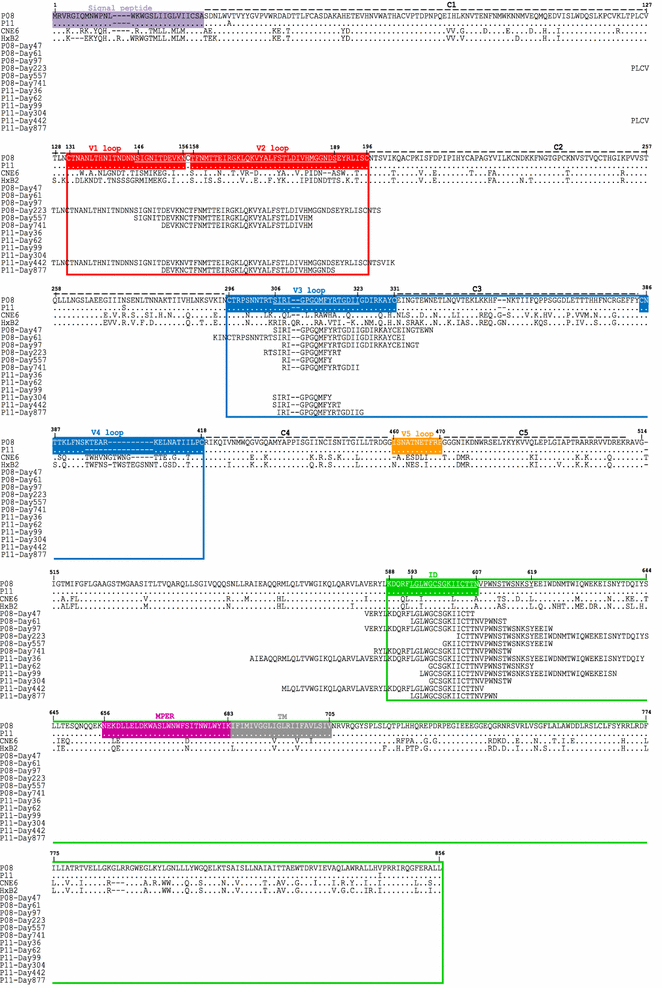
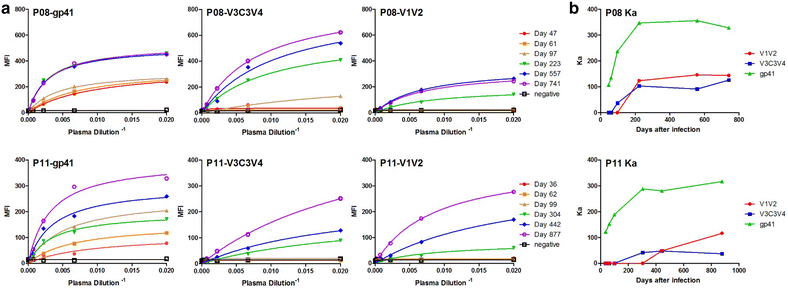
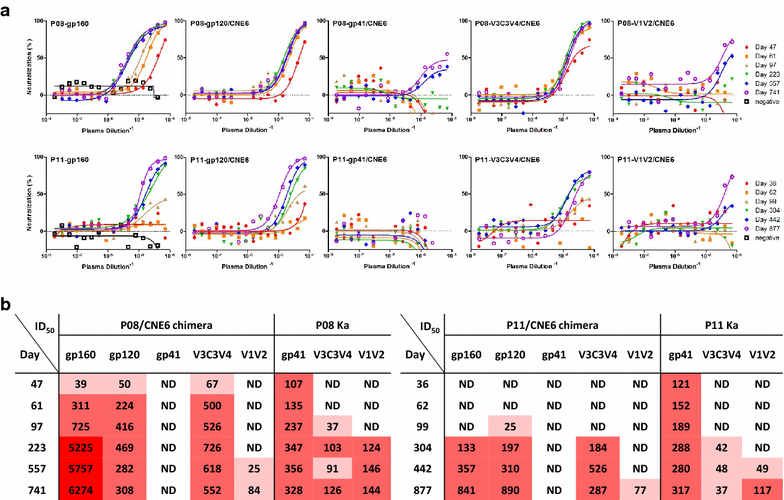
Results: We report here antibody profiling of sequential plasma samples against transmitted/founder HIV-1 envelope glycoprotein in an epidemiologically linked transmission pair using our previously reported antigen library approach. We have decomposed the antibody recognition into three major subdomains on the envelope and showed their development in vivo followed a spatiotemporal hierarchy: starting with the ectodomain of gp41 at membrane proximal region, then the V3C3V4 and the V1V2 of gp120 at the membrane distal region. While antibodies to these subdomains appeared to undergo avidity maturation, the early anti-gp41 antibodies failed to translate into detectable autologous neutralization. Instead, it was the much delayed anti-V3C3V4 and anti-V1V2 antibodies constituted the major neutralizing activities.
Conclusions: Our results indicate that the initial antibody response was severely misguided by the transmitted/founder virus and future vaccine design would need to avoid the ectodomain of gp41 and focus on the neutralizing targets in the V3C3V4 and V1V2 subdomains of gp120.
Figures

Similar articles
-
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.J Virol. 2016 Jan 13;90(7):3446-57. doi: 10.1128/JVI.03090-15. J Virol. 2016. PMID: 26763999 Free PMC article.
-
Rationally Targeted Mutations at the V1V2 Domain of the HIV-1 Envelope to Augment Virus Neutralization by Anti-V1V2 Monoclonal Antibodies.PLoS One. 2015 Oct 22;10(10):e0141233. doi: 10.1371/journal.pone.0141233. eCollection 2015. PLoS One. 2015. PMID: 26491873 Free PMC article.
-
Immunogenicity of a Prefusion HIV-1 Envelope Trimer in Complex with a Quaternary-Structure-Specific Antibody.J Virol. 2015 Dec 30;90(6):2740-55. doi: 10.1128/JVI.02380-15. J Virol. 2015. PMID: 26719262 Free PMC article.
-
HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies.Expert Rev Vaccines. 2016;15(3):349-65. doi: 10.1586/14760584.2016.1129905. Epub 2016 Jan 8. Expert Rev Vaccines. 2016. PMID: 26654478 Review.
-
Specificity of the autologous neutralizing antibody response.Curr Opin HIV AIDS. 2009 Sep;4(5):358-63. doi: 10.1097/COH.0b013e32832ea7e8. Curr Opin HIV AIDS. 2009. PMID: 20048698 Free PMC article. Review.
Cited by
-
Higher HIV-1 Env gp120-Specific Antibody-Dependent Cellular Cytotoxicity (ADCC) Activity Is Associated with Lower Levels of Defective HIV-1 Provirus.Viruses. 2023 Oct 6;15(10):2055. doi: 10.3390/v15102055. Viruses. 2023. PMID: 37896832 Free PMC article.
-
Epitope-focused immunogens against the CD4-binding site of HIV-1 envelope protein induce neutralizing antibodies against auto- and heterologous viruses.J Biol Chem. 2018 Jan 19;293(3):830-846. doi: 10.1074/jbc.M117.816447. Epub 2017 Nov 29. J Biol Chem. 2018. PMID: 29187598 Free PMC article.
-
Conformational plasticity of the HIV-1 gp41 immunodominant region is recognized by multiple non-neutralizing antibodies.Commun Biol. 2022 Mar 31;5(1):291. doi: 10.1038/s42003-022-03235-w. Commun Biol. 2022. PMID: 35361878 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

