S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification
- PMID: 26883378
- PMCID: PMC7172980
- DOI: 10.1016/j.meegid.2016.02.015
S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification
Abstract
Infectious bronchitis virus (IBV) is the causative agent of a highly contagious disease that results in severe economic losses to the global poultry industry. The virus exists in a wide variety of genetically distinct viral types, and both phylogenetic analysis and measures of pairwise similarity among nucleotide or amino acid sequences have been used to classify IBV strains. However, there is currently no consensus on the method by which IBV sequences should be compared, and heterogeneous genetic group designations that are inconsistent with phylogenetic history have been adopted, leading to the confusing coexistence of multiple genotyping schemes. Herein, we propose a simple and repeatable phylogeny-based classification system combined with an unambiguous and rationale lineage nomenclature for the assignment of IBV strains. By using complete nucleotide sequences of the S1 gene we determined the phylogenetic structure of IBV, which in turn allowed us to define 6 genotypes that together comprise 32 distinct viral lineages and a number of inter-lineage recombinants. Because of extensive rate variation among IBVs, we suggest that the inference of phylogenetic relationships alone represents a more appropriate criterion for sequence classification than pairwise sequence comparisons. The adoption of an internationally accepted viral nomenclature is crucial for future studies of IBV epidemiology and evolution, and the classification scheme presented here can be updated and revised novel S1 sequences should become available.
Keywords: Classification; Evolution; IBV; Phylogeny.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures
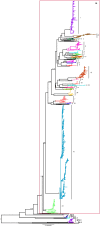
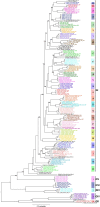
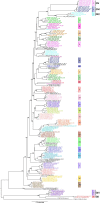
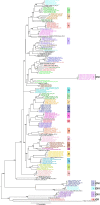
References
-
- Abdel-Moneim A.S., Madbouly H.M., Gelb J., Jr., Ladman B.S. Isolation and identification of Egypt/Beni-Suef/01 a novel genotype of infectious bronchitis virus. Vet. Med. J. Giza. 2002;50:1065–1078.
-
- Abdel-Moneim A.S., Afifi M.A., El-Kady M.F. Emergence of a novel genotype of avian infectious bronchitis virus in Egypt. Arch. Virol. 2012;157(12):2453–2457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

