Great ape Y Chromosome and mitochondrial DNA phylogenies reflect subspecies structure and patterns of mating and dispersal
- PMID: 26883546
- PMCID: PMC4817767
- DOI: 10.1101/gr.198754.115
Great ape Y Chromosome and mitochondrial DNA phylogenies reflect subspecies structure and patterns of mating and dispersal
Abstract
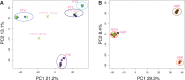
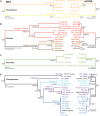
The distribution of genetic diversity in great ape species is likely to have been affected by patterns of dispersal and mating. This has previously been investigated by sequencing autosomal and mitochondrial DNA (mtDNA), but large-scale sequence analysis of the male-specific region of the Y Chromosome (MSY) has not yet been undertaken. Here, we use the human MSY reference sequence as a basis for sequence capture and read mapping in 19 great ape males, combining the data with sequences extracted from the published whole genomes of 24 additional males to yield a total sample of 19 chimpanzees, four bonobos, 14 gorillas, and six orangutans, in which interpretable MSY sequence ranges from 2.61 to 3.80 Mb. This analysis reveals thousands of novel MSY variants and defines unbiased phylogenies. We compare these with mtDNA-based trees in the same individuals, estimating time-to-most-recent common ancestor (TMRCA) for key nodes in both cases. The two loci show high topological concordance and are consistent with accepted (sub)species definitions, but time depths differ enormously between loci and (sub)species, likely reflecting different dispersal and mating patterns. Gorillas and chimpanzees/bonobos present generally low and high MSY diversity, respectively, reflecting polygyny versus multimale-multifemale mating. However, particularly marked differences exist among chimpanzee subspecies: The western chimpanzee MSY phylogeny has a TMRCA of only 13.2 (10.8-15.8) thousand years, but that for central chimpanzees exceeds 1 million years. Cross-species comparison within a single MSY phylogeny emphasizes the low human diversity, and reveals species-specific branch length variation that may reflect differences in long-term generation times.
© 2016 Hallast et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Archidiacono N, Storlazzi CT, Spalluto C, Ricco AS, Marzella R, Rocchi M. 1998. Evolution of chromosome Y in primates. Chromosoma 107: 241–246. - PubMed
-
- Batini C, Lopes J, Behar DM, Calafell F, Jorde LB, van der Veen L, Quintana-Murci L, Spedini G, Destro-Bisol G, Comas D. 2011. Insights into the demographic history of African Pygmies from complete mitochondrial genomes. Mol Biol Evol 28: 1099–1110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
