Deciphering the importance of the palindromic architecture of the immunoglobulin heavy-chain 3' regulatory region
- PMID: 26883548
- PMCID: PMC4757795
- DOI: 10.1038/ncomms10730
Deciphering the importance of the palindromic architecture of the immunoglobulin heavy-chain 3' regulatory region
Abstract
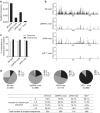
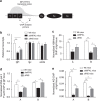
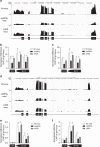
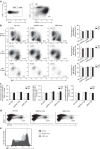
The IgH 3' regulatory region (3'RR) controls class switch recombination (CSR) and somatic hypermutation (SHM) in B cells. The mouse 3'RR contains four enhancer elements with hs1,2 flanked by inverted repeated sequences and the centre of a 25-kb palindrome bounded by two hs3 enhancer inverted copies (hs3a and hs3b). hs4 lies downstream of the palindrome. In mammals, evolution maintained this unique palindromic arrangement, suggesting that it is functionally significant. Here we report that deconstructing the palindromic IgH 3'RR strongly affects its function even when enhancers are preserved. CSR and IgH transcription appear to be poorly dependent on the 3'RR architecture and it is more or less preserved, provided 3'RR enhancers are present. By contrast, a 'palindromic effect' significantly lowers VH germline transcription, AID recruitment and SHM. In conclusion, this work indicates that the IgH 3'RR does not simply pile up enhancer units but also optimally exposes them into a functional architecture of crucial importance.
Figures




References
-
- Henderson A. & Calame K. Transcription regulation during B cell development. Annu. Rev. Immunol. 16, 163–200 (1998). - PubMed
-
- Pinaud E. et al.. The IgH locus 3' regulatory region: pulling the strings from behind. Adv. Immunol. 110, 27–70 (2011). - PubMed
-
- Marquet M. et al.. The Eμ enhancer region influences H chain expression and B cell fate without impacting IgVH repertoire and immune response in vivo. J. Immunol. 193, 1171–1183 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

