A SUMO-acetyl switch in PXR biology
- PMID: 26883953
- PMCID: PMC4975675
- DOI: 10.1016/j.bbagrm.2016.02.008
A SUMO-acetyl switch in PXR biology
Abstract
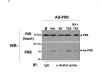
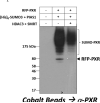
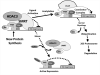
Post-translational modification (PTM) of nuclear receptor superfamily members regulates various aspects of their biology to include sub-cellular localization, the repertoire of protein-binding partners, as well as their stability and mode of degradation. The nuclear receptor pregnane X receptor (PXR, NR1I2) is a master-regulator of the drug-inducible gene expression in liver and intestine. The PXR-mediated gene activation program is primarily recognized to increase drug metabolism, drug transport, and drug efflux pathways in these tissues. The activation of PXR also has important implications in significant human diseases including inflammatory bowel disease and cancer. Our recent investigations reveal that PXR is modified by multiple PTMs to include phosphorylation, SUMOylation, and ubiquitination. Using both primary cultures of hepatocytes and cell-based assays, we show here that PXR is modified through acetylation on lysine residues. Further, we show that increased acetylation of PXR stimulates its increased SUMO-modification to support active transcriptional suppression. Pharmacologic inhibition of lysine de-acetylation using trichostatin A (TSA) alters the sub-cellular localization of PXR in cultured hepatocytes, and also has a profound impact upon PXR transactivation capacity. Both the acetylation and SUMOylation status of the PXR protein is affected by its ability to associate with the lysine de-acetylating enzyme histone de-acetylase (HDAC)3 in a complex with silencing mediator of retinoic acid and thyroid hormone receptor (SMRT). Taken together, our data support a model in which a SUMO-acetyl 'switch' occurs such that acetylation of PXR likely stimulates SUMO-modification of PXR to promote the active repression of PXR-target gene expression. This article is part of a Special Issue entitled: Xenobiotic nuclear receptors: New Tricks for An Old Dog, edited by Dr. Wen Xie.
Keywords: Acetylation; Histone deacetylase 3; Nuclear receptor; Post-translational modification; Pregnane X receptor; SUMOylation; Silencing mediator of retinoic acid and thyroid hormone receptor; Ubiquitination.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures
















Similar articles
-
Acetylation of lysine 109 modulates pregnane X receptor DNA binding and transcriptional activity.Biochim Biophys Acta. 2016 Sep;1859(9):1155-1169. doi: 10.1016/j.bbagrm.2016.01.006. Epub 2016 Feb 23. Biochim Biophys Acta. 2016. PMID: 26855179 Free PMC article.
-
SUMOylation and Ubiquitylation Circuitry Controls Pregnane X Receptor Biology in Hepatocytes.Drug Metab Dispos. 2015 Sep;43(9):1316-25. doi: 10.1124/dmd.115.065201. Epub 2015 Jun 10. Drug Metab Dispos. 2015. PMID: 26063058 Free PMC article.
-
Genomewide comparison of the inducible transcriptomes of nuclear receptors CAR, PXR and PPARα in primary human hepatocytes.Biochim Biophys Acta. 2016 Sep;1859(9):1218-1227. doi: 10.1016/j.bbagrm.2016.03.007. Epub 2016 Mar 17. Biochim Biophys Acta. 2016. PMID: 26994748
-
Epigenetic regulation of pregnane X receptor activity.Drug Metab Rev. 2013 May;45(2):166-72. doi: 10.3109/03602532.2012.756012. Drug Metab Rev. 2013. PMID: 23600685 Review.
-
Post-translational modification of pregnane x receptor.Pharmacol Res. 2011 Jul;64(1):4-10. doi: 10.1016/j.phrs.2011.02.011. Epub 2011 Mar 21. Pharmacol Res. 2011. PMID: 21397695 Free PMC article. Review.
Cited by
-
The pregnane X receptor drives sexually dimorphic hepatic changes in lipid and xenobiotic metabolism in response to gut microbiota in mice.Microbiome. 2021 Apr 20;9(1):93. doi: 10.1186/s40168-021-01050-9. Microbiome. 2021. PMID: 33879258 Free PMC article.
-
Regulation of PXR Function by Coactivator and Corepressor Proteins: Ligand Binding Is Just the Beginning.Cells. 2021 Nov 12;10(11):3137. doi: 10.3390/cells10113137. Cells. 2021. PMID: 34831358 Free PMC article. Review.
-
Novel complex of HAT protein TIP60 and nuclear receptor PXR promotes cell migration and adhesion.Sci Rep. 2017 Jun 16;7(1):3635. doi: 10.1038/s41598-017-03783-w. Sci Rep. 2017. PMID: 28623334 Free PMC article.
-
The Roles of SUMO in Metabolic Regulation.Adv Exp Med Biol. 2017;963:143-168. doi: 10.1007/978-3-319-50044-7_9. Adv Exp Med Biol. 2017. PMID: 28197911 Free PMC article. Review.
-
Nuclear Receptor PXR Confers Irradiation Resistance by Promoting DNA Damage Response Through Stabilization of ATF3.Front Oncol. 2022 Mar 16;12:837980. doi: 10.3389/fonc.2022.837980. eCollection 2022. Front Oncol. 2022. PMID: 35372071 Free PMC article.
References
-
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

