LEAFDATA: a literature-curated database for Arabidopsis leaf development
- PMID: 26884807
- PMCID: PMC4754890
- DOI: 10.1186/s13007-016-0115-9
LEAFDATA: a literature-curated database for Arabidopsis leaf development
Abstract
Background: In the post-genomic era, biological databases provide an easy access to a wide variety of scientific data. The vast quantity of literature calls for curated databases where existing knowledge is carefully organized in order to aid novel discoveries. Leaves, the main photosynthetic organs are not only vital for plant growth but also essential for maintaining the global ecosystem by producing oxygen and food. Therefore, studying and understanding leaf formation and growth are key objectives in biology. Arabidopsis thaliana to this date remains the prime experimental model organism in plant science.
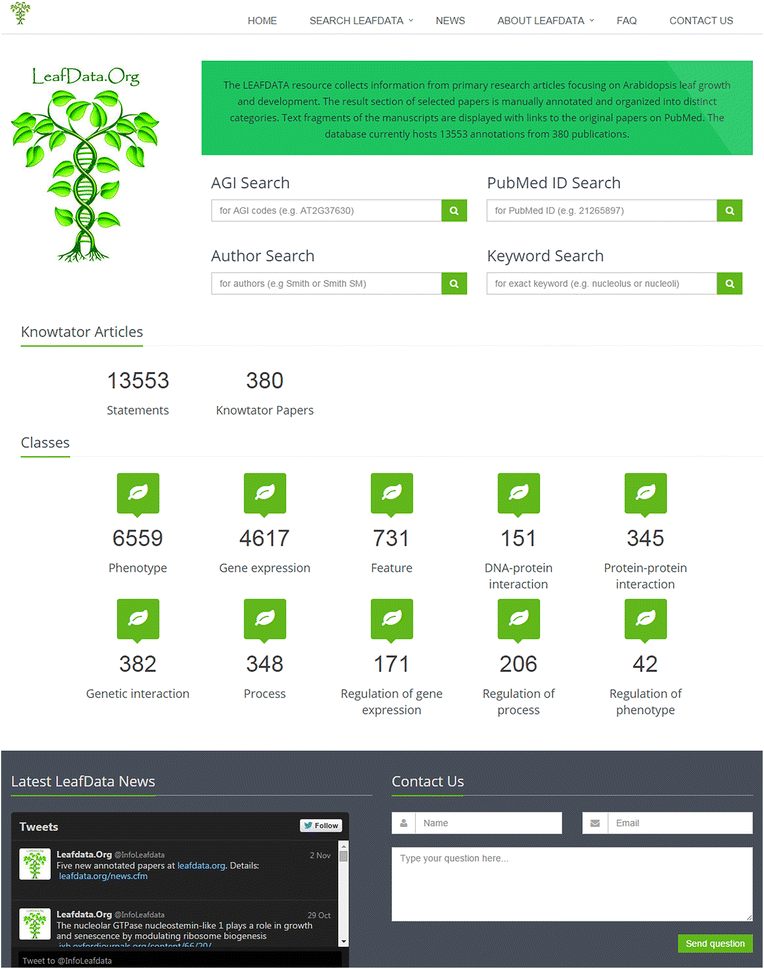
Description: LEAFDATA was created as an easily accessible and searchable web tool to assemble a relevant collection of Arabidopsis leaf literature. LEAFDATA currently contains 13,553 categorized statements from 380 processed publications. LEAFDATA can be searched for genes of interest using Arabidopsis Genome Initiative identifiers, for selected papers by means of PubMed IDs, authors and specific keywords. The results page contains details of the original publications, text fragments from the curated literature grouped according to information types and direct links to PubMed pages of the original papers.
Conclusions: The LEAFDATA database offers access to searchable entries curated from a large number of scientific publications. Due to the unprecedented details of annotations and the fact that LEAFDATA already provides records about approximately 1600 individual loci, this database is useful for the entire plant research community.
Keywords: Arabidopsis thaliana; Database; Leaf development; Manual literature curation.
Figures

Similar articles
-
PlantMP: a database for moonlighting plant proteins.Database (Oxford). 2019 Jan 1;2019:baz050. doi: 10.1093/database/baz050. Database (Oxford). 2019. PMID: 31032837 Free PMC article.
-
TMDB: a literature-curated database for small molecular compounds found from tea.BMC Plant Biol. 2014 Sep 16;14:243. doi: 10.1186/s12870-014-0243-1. BMC Plant Biol. 2014. PMID: 25224438 Free PMC article.
-
A framework for community curation of interspecies interactions literature.Elife. 2023 Jul 4;12:e84658. doi: 10.7554/eLife.84658. Elife. 2023. PMID: 37401199 Free PMC article.
-
The Alliance of Genome Resources: Building a Modern Data Ecosystem for Model Organism Databases.Genetics. 2019 Dec;213(4):1189-1196. doi: 10.1534/genetics.119.302523. Genetics. 2019. PMID: 31796553 Free PMC article. Review.
-
DBHR: a collection of databases relevant to human research.Future Sci OA. 2021 Jan 20;8(3):FSO780. doi: 10.2144/fsoa-2021-0101. eCollection 2022 Mar. Future Sci OA. 2021. PMID: 35251694 Free PMC article. Review.
Cited by
-
Identification and characterization of ncRNA-associated ceRNA networks in Arabidopsis leaf development.BMC Genomics. 2018 Aug 13;19(1):607. doi: 10.1186/s12864-018-4993-2. BMC Genomics. 2018. PMID: 30103673 Free PMC article.
-
The Gastrodia menghaiensis (Orchidaceae) genome provides new insights of orchid mycorrhizal interactions.BMC Plant Biol. 2022 Apr 7;22(1):179. doi: 10.1186/s12870-022-03573-1. BMC Plant Biol. 2022. PMID: 35392808 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

