The mid-developmental transition and the evolution of animal body plans
- PMID: 26886793
- PMCID: PMC4817236
- DOI: 10.1038/nature16994
The mid-developmental transition and the evolution of animal body plans
Erratum in
-
Author Correction: The mid-developmental transition and the evolution of animal body plans.Nature. 2019 Nov;575(7782):E3. doi: 10.1038/s41586-019-1698-x. Nature. 2019. PMID: 31673121
Abstract
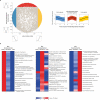
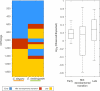
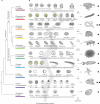
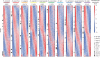
Animals are grouped into ~35 'phyla' based upon the notion of distinct body plans. Morphological and molecular analyses have revealed that a stage in the middle of development--known as the phylotypic period--is conserved among species within some phyla. Although these analyses provide evidence for their existence, phyla have also been criticized as lacking an objective definition, and consequently based on arbitrary groupings of animals. Here we compare the developmental transcriptomes of ten species, each annotated to a different phylum, with a wide range of life histories and embryonic forms. We find that in all ten species, development comprises the coupling of early and late phases of conserved gene expression. These phases are linked by a divergent 'mid-developmental transition' that uses species-specific suites of signalling pathways and transcription factors. This mid-developmental transition overlaps with the phylotypic period that has been defined previously for three of the ten phyla, suggesting that transcriptional circuits and signalling mechanisms active during this transition are crucial for defining the phyletic body plan and that the mid-developmental transition may be used to define phylotypic periods in other phyla. Placing these observations alongside the reported conservation of mid-development within phyla, we propose that a phylum may be defined as a collection of species whose gene expression at the mid-developmental transition is both highly conserved among them, yet divergent relative to other species.
Figures
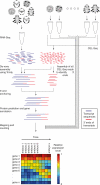
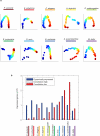
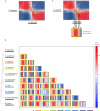
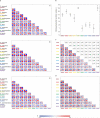


Comment in
-
Animal Evolution: Are Phyla Real?Curr Biol. 2016 May 23;26(10):R424-6. doi: 10.1016/j.cub.2016.03.058. Curr Biol. 2016. PMID: 27218852
References
-
- Valentine JW. On the Origin of Phyla. Univ. Chicago Press; 2004.
-
- Haeckel E. The Evolution of Man. Vol. 1. C. K. Paul & Company; 1879.
-
- Gould SJ. Ontogeny and Phylogeny. Harvard Univ. Press; 1977.
-
- Abzhanov A. Von Baer's law for the ages: lost and found principles of developmental evolution. Trends Genet. 2013;29:712–722. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

