CellECT: cell evolution capturing tool
- PMID: 26887436
- PMCID: PMC4756481
- DOI: 10.1186/s12859-016-0927-7
CellECT: cell evolution capturing tool
Abstract
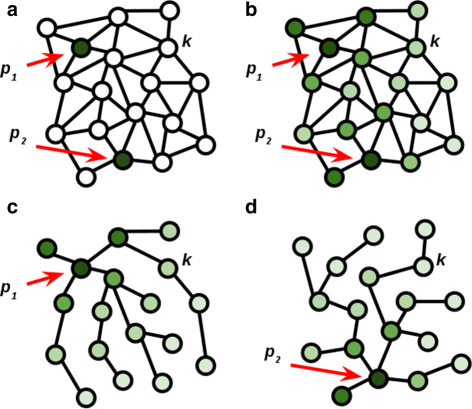
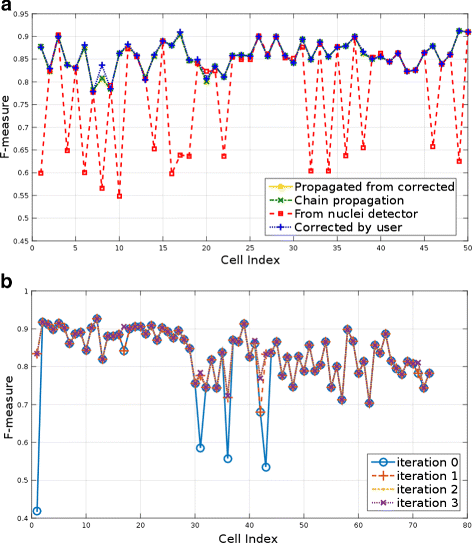
Background: Robust methods for the segmentation and analysis of cells in 3D time sequences (3D+t) are critical for quantitative cell biology. While many automated methods for segmentation perform very well, few generalize reliably to diverse datasets. Such automated methods could significantly benefit from at least minimal user guidance. Identification and correction of segmentation errors in time-series data is of prime importance for proper validation of the subsequent analysis. The primary contribution of this work is a novel method for interactive segmentation and analysis of microscopy data, which learns from and guides user interactions to improve overall segmentation.
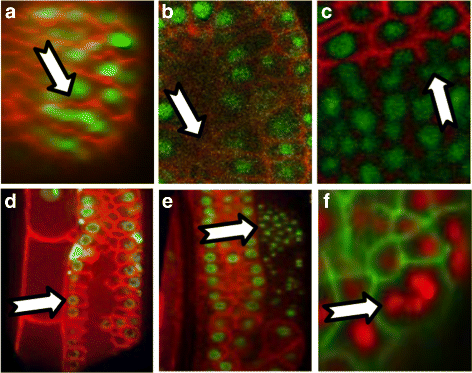
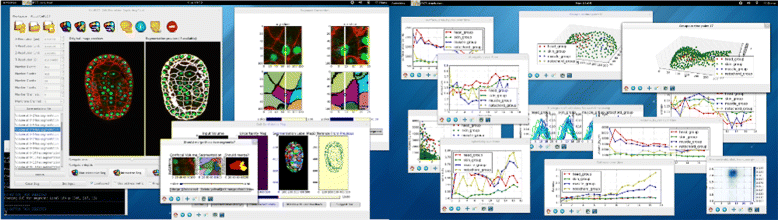
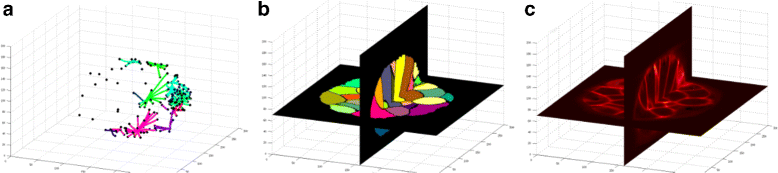
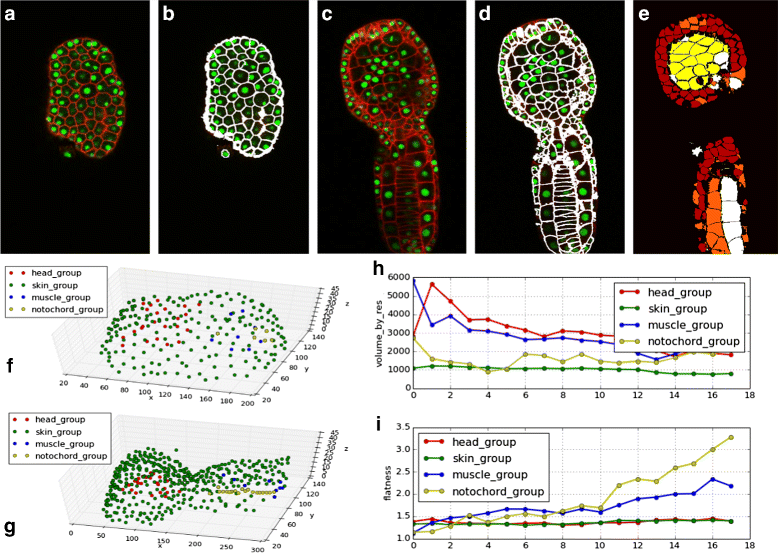
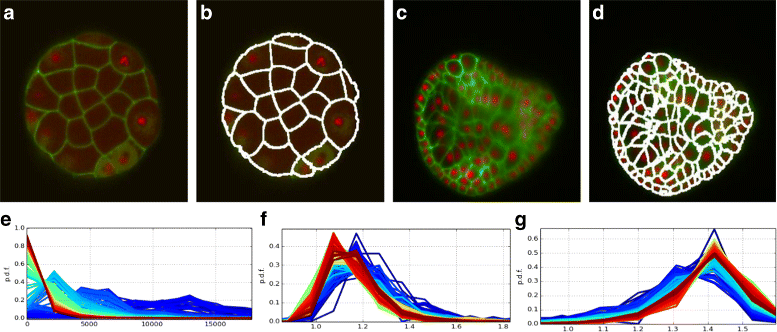
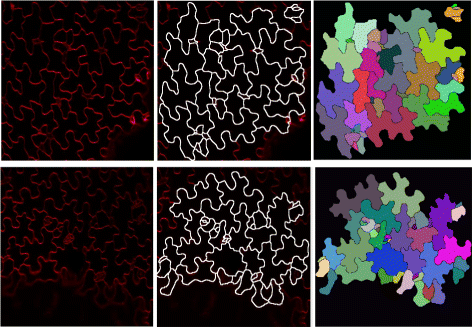
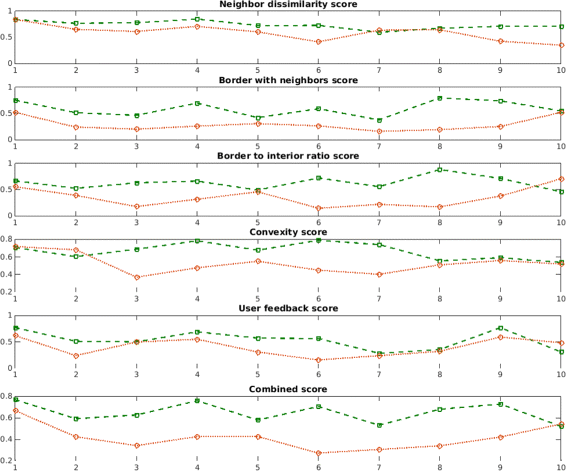
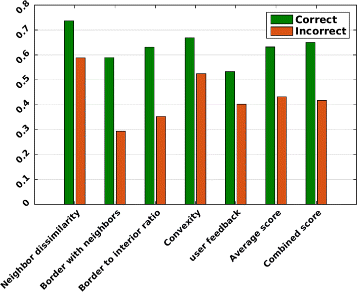
Results: We introduce an interactive cell analysis application, called CellECT, for 3D+t microscopy datasets. The core segmentation tool is watershed-based and allows the user to add, remove or modify existing segments by means of manipulating guidance markers. A confidence metric learns from the user interaction and highlights regions of uncertainty in the segmentation for the user's attention. User corrected segmentations are then propagated to neighboring time points. The analysis tool computes local and global statistics for various cell measurements over the time sequence. Detailed results on two large datasets containing membrane and nuclei data are presented: a 3D+t confocal microscopy dataset of the ascidian Phallusia mammillata consisting of 18 time points, and a 3D+t single plane illumination microscopy (SPIM) dataset consisting of 192 time points. Additionally, CellECT was used to segment a large population of jigsaw-puzzle shaped epidermal cells from Arabidopsis thaliana leaves. The cell coordinates obtained using CellECT are compared to those of manually segmented cells.
Conclusions: CellECT provides tools for convenient segmentation and analysis of 3D+t membrane datasets by incorporating human interaction into automated algorithms. Users can modify segmentation results through the help of guidance markers, and an adaptive confidence metric highlights problematic regions. Segmentations can be propagated to multiple time points, and once a segmentation is available for a time sequence cells can be analyzed to observe trends. The segmentation and analysis tools presented here generalize well to membrane or cell wall volumetric time series datasets.
Figures



References
-
- Derde M, Antanas L, De Raedt L, Guiza Grandas F. An interactive learning approach to histology image segmentation. In: Proceedings of the 24th Benelux Conference on Artificial Intelligence: 2012. p. 1–8.
-
- Sommer C, Straehle C, Kothe U, Hamprecht FA. Ilastik: Interactive learning and segmentation toolkit. In: Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium On. IEEE: 2011. p. 230–233.
-
- McGuinness K, O’Connor NE. A comparative evaluation of interactive segmentation algorithms. Pattern Recognit. 2010;43(2):434–44. doi: 10.1016/j.patcog.2009.03.008. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

