A Modular Assembly Platform for Rapid Generation of DNA Constructs
- PMID: 26887506
- PMCID: PMC4757859
- DOI: 10.1038/srep16836
A Modular Assembly Platform for Rapid Generation of DNA Constructs
Abstract
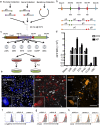
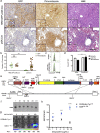
Traditional cloning methods have limitations on the number of DNA fragments that can be simultaneously manipulated, which dramatically slows the pace of molecular assembly. Here we describe GMAP, a Gibson assembly-based modular assembly platform consisting of a collection of promoters and genes, which allows for one-step production of DNA constructs. GMAP facilitates rapid assembly of expression and viral constructs using modular genetic components, as well as increasingly complicated genetic tools using contextually relevant genomic elements. Our data demonstrate the applicability of GMAP toward the validation of synthetic promoters, identification of potent RNAi constructs, establishment of inducible lentiviral systems, tumor initiation in genetically engineered mouse models, and gene-targeting for the generation of knock-in mice. GMAP represents a recombinant DNA technology designed for widespread circulation and easy adaptation for other uses, such as synthetic biology, genetic screens, and CRISPR-Cas9.
Figures
References
-
- Lodish H. et al. Molecular Cell Biology. (Freeman W. H., 2000).
-
- Li M. Z. & Elledge S. J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007). - PubMed
-
- Werner S., Engler C., Weber E., Gruetzner R. & Marillonnet S. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng. Bugs 3, 38–43 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

