Classification of low quality cells from single-cell RNA-seq data
- PMID: 26887813
- PMCID: PMC4758103
- DOI: 10.1186/s13059-016-0888-1
Classification of low quality cells from single-cell RNA-seq data
Abstract
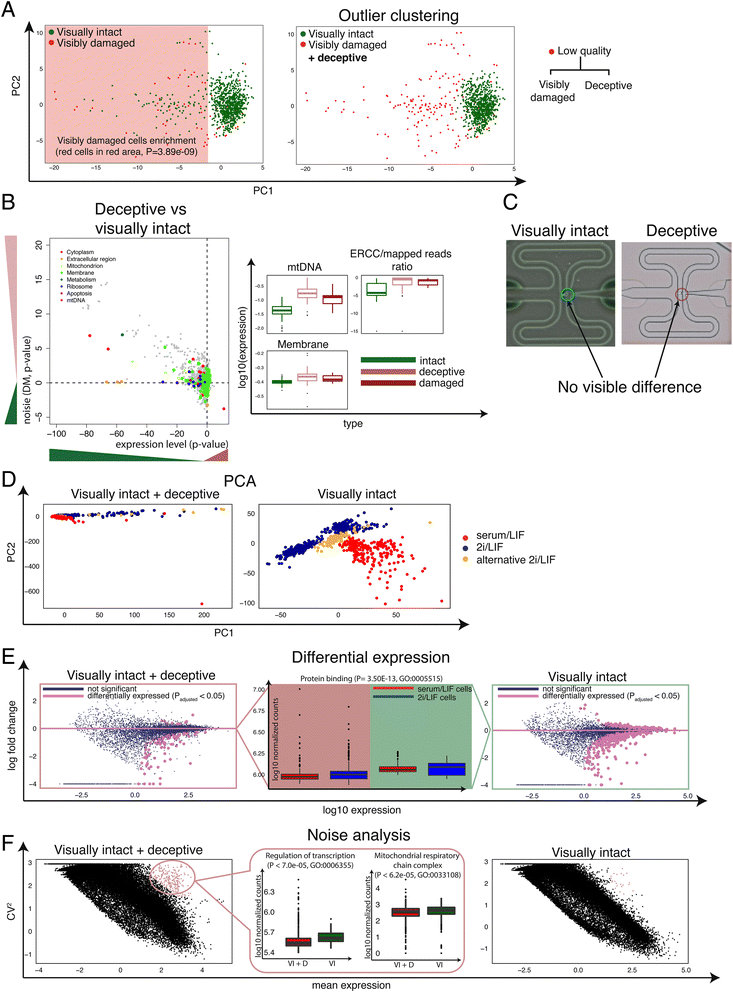
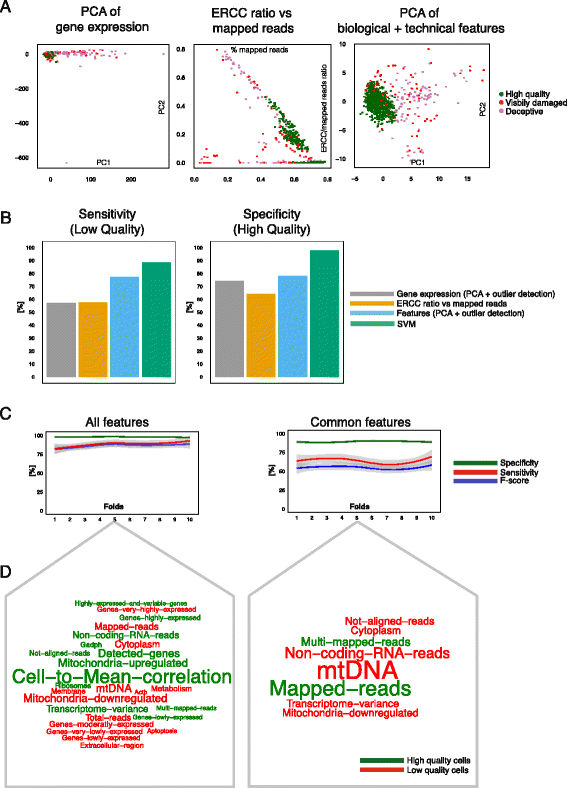
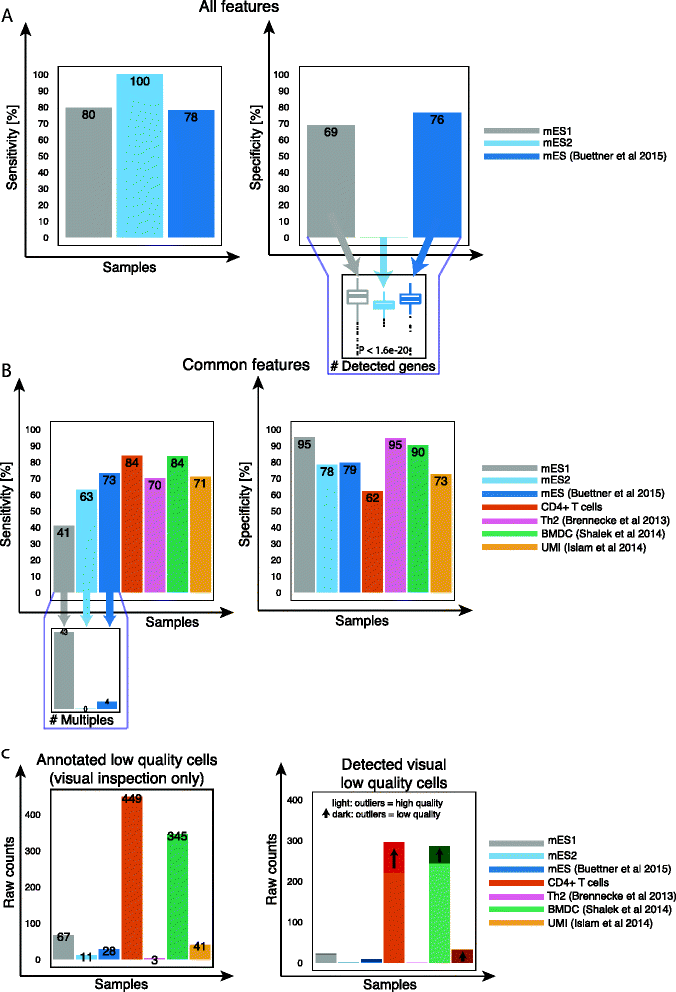
Single-cell RNA sequencing (scRNA-seq) has broad applications across biomedical research. One of the key challenges is to ensure that only single, live cells are included in downstream analysis, as the inclusion of compromised cells inevitably affects data interpretation. Here, we present a generic approach for processing scRNA-seq data and detecting low quality cells, using a curated set of over 20 biological and technical features. Our approach improves classification accuracy by over 30 % compared to traditional methods when tested on over 5,000 cells, including CD4+ T cells, bone marrow dendritic cells, and mouse embryonic stem cells.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

