Differentiated, Promoter-specific Response of [4Fe-4S] NsrR DNA Binding to Reaction with Nitric Oxide
- PMID: 26887943
- PMCID: PMC4861436
- DOI: 10.1074/jbc.M115.693192
Differentiated, Promoter-specific Response of [4Fe-4S] NsrR DNA Binding to Reaction with Nitric Oxide
Abstract
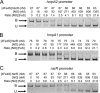
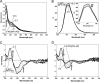
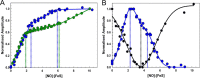
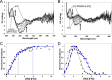
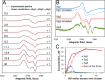
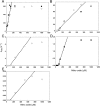
NsrR is an iron-sulfur cluster protein that regulates the nitric oxide (NO) stress response of many bacteria. NsrR from Streptomyces coelicolor regulates its own expression and that of only two other genes, hmpA1 and hmpA2, which encode HmpA enzymes predicted to detoxify NO. NsrR binds promoter DNA with high affinity only when coordinating a [4Fe-4S] cluster. Here we show that reaction of [4Fe-4S] NsrR with NO affects DNA binding differently depending on the gene promoter. Binding to the hmpA2 promoter was abolished at ∼2 NO per cluster, although for the hmpA1 and nsrR promoters, ∼4 and ∼8 NO molecules, respectively, were required to abolish DNA binding. Spectroscopic and kinetic studies of the NO reaction revealed a rapid, multi-phase, non-concerted process involving up to 8-10 NO molecules per cluster, leading to the formation of several iron-nitrosyl species. A distinct intermediate was observed at ∼2 NO per cluster, along with two further intermediates at ∼4 and ∼6 NO. The NsrR nitrosylation reaction was not significantly affected by DNA binding. These results show that NsrR regulates different promoters in response to different concentrations of NO. Spectroscopic evidence indicates that this is achieved by different NO-FeS complexes.
Keywords: DNA-binding protein; DNA-protein interaction; iron; iron-sulfur protein; nitric oxide; regulator; spectroscopy.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
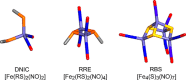

References
-
- Crack J. C., Green J., Thomson A. J., and Le Brun N. E. (2012) Iron-sulfur cluster sensor-regulators. Curr. Opin. Chem. Biol. 16, 35–44 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

