Genomic and transcriptomic comparison of nucleotide variations for insights into bruchid resistance of mungbean (Vigna radiata [L.] R. Wilczek)
- PMID: 26887961
- PMCID: PMC4756517
- DOI: 10.1186/s12870-016-0736-1
Genomic and transcriptomic comparison of nucleotide variations for insights into bruchid resistance of mungbean (Vigna radiata [L.] R. Wilczek)
Abstract
Background: Mungbean (Vigna radiata [L.] R. Wilczek) is an important legume crop with high nutritional value in South and Southeast Asia. The crop plant is susceptible to a storage pest caused by bruchids (Callosobruchus spp.). Some wild and cultivated mungbean accessions show resistance to bruchids. Genomic and transcriptomic comparison of bruchid-resistant and -susceptible mungbean could reveal bruchid-resistant genes (Br) for this pest and give insights into the bruchid resistance of mungbean.
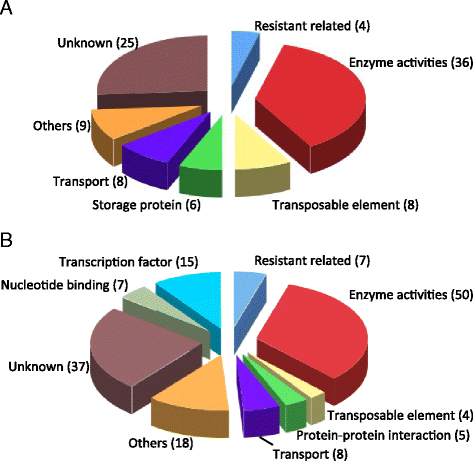
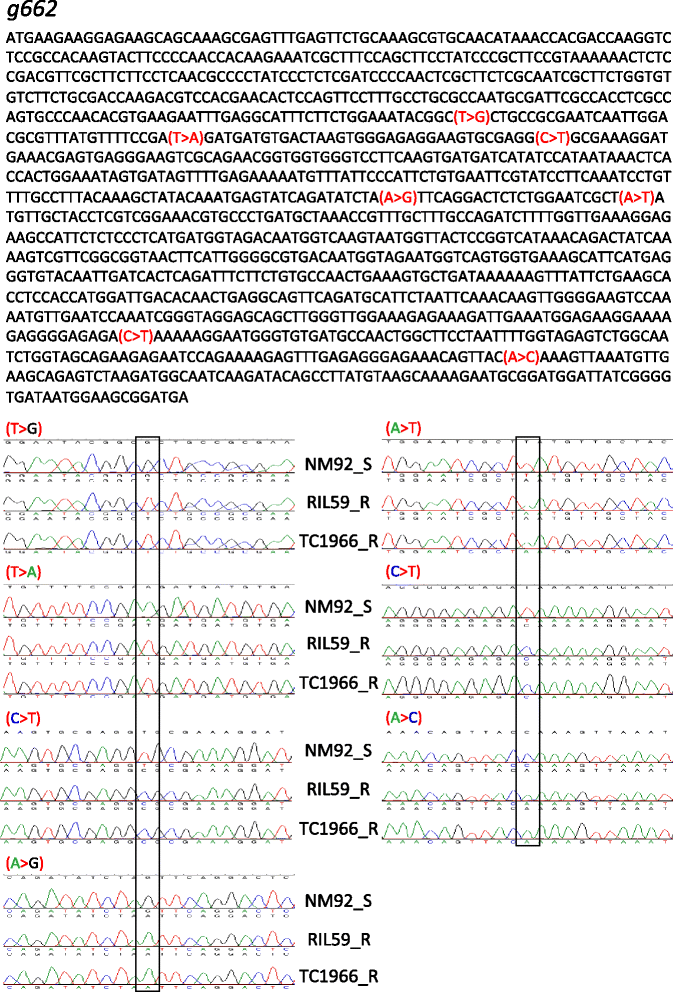
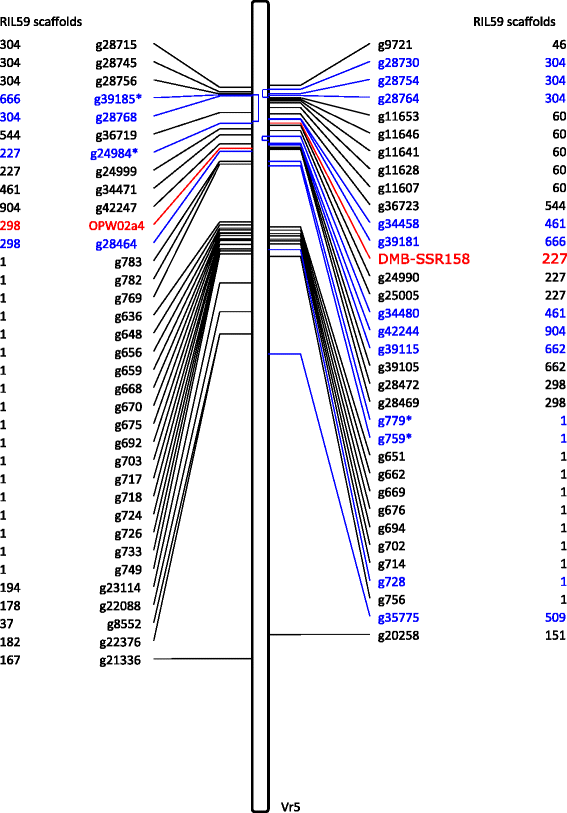
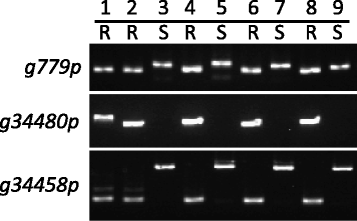
Results: Flow cytometry showed that the genome size varied by 61 Mb (mega base pairs) among the tested mungbean accessions. Next generation sequencing followed by de novo assembly of the genome of the bruchid-resistant recombinant inbred line 59 (RIL59) revealed more than 42,000 genes. Transcriptomic comparison of bruchid-resistant and -susceptible parental lines and their offspring identified 91 differentially expressed genes (DEGs) classified into 17 major and 74 minor bruchid-resistance-associated genes. We found 408 nucleotide variations (NVs) between bruchid-resistant and -susceptible lines in regions spanning 2 kb (kilo base pairs) of the promoters of 68 DEGs. Furthermore, 282 NVs were identified on exons of 148 sequence-changed-protein genes (SCPs). DEGs and SCPs comprised genes involved in resistant-related, transposable elements (TEs) and conserved metabolic pathways. A large number of these genes were mapped to a region on chromosome 5. Molecular markers designed for variants of putative bruchid-resistance-associated genes were highly diagnostic for the bruchid-resistant genotype.
Conclusions: In addition to identifying bruchid-resistance-associated genes, we found that conserved metabolism and TEs may be modifier factors for bruchid resistance of mungbean. The genome sequence of a bruchid-resistant inbred line, candidate genes and sequence variations in promoter regions and exons putatively conditioning resistance as well as markers detecting these variants could be used for development of bruchid-resistant mungbean varieties.
Figures



Similar articles
-
Mechanism of Resistance in Mungbean [Vigna radiata (L.) R. Wilczek var. radiata] to bruchids, Callosobruchus spp. (Coleoptera: Bruchidae).Front Plant Sci. 2017 Jun 20;8:1031. doi: 10.3389/fpls.2017.01031. eCollection 2017. Front Plant Sci. 2017. PMID: 28676807 Free PMC article. Review.
-
Transcriptomic and Proteomic Research To Explore Bruchid-Resistant Genes in Mungbean Isogenic Lines.J Agric Food Chem. 2016 Aug 31;64(34):6648-58. doi: 10.1021/acs.jafc.6b03015. Epub 2016 Aug 17. J Agric Food Chem. 2016. PMID: 27508985
-
Identification of single nucleotide polymorphism markers associated with resistance to bruchids (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) and cultivated V. radiata through genotyping by sequencing and quantitative trait locus analysis.BMC Plant Biol. 2016 Jul 15;16(1):159. doi: 10.1186/s12870-016-0847-8. BMC Plant Biol. 2016. PMID: 27422285 Free PMC article.
-
A second VrPGIP1 allele is associated with bruchid resistance (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) accession ACC41.Mol Genet Genomics. 2020 Mar;295(2):275-286. doi: 10.1007/s00438-019-01619-y. Epub 2019 Nov 8. Mol Genet Genomics. 2020. PMID: 31705195
-
Genetic analysis and identification of VrFRO8, a salt tolerance-related gene in mungbean.Gene. 2022 Aug 20;836:146658. doi: 10.1016/j.gene.2022.146658. Epub 2022 Jun 14. Gene. 2022. PMID: 35714797 Review.
Cited by
-
Chilling susceptibility in mungbean varieties is associated with their differentially expressed genes.Bot Stud. 2017 Dec;58(1):7. doi: 10.1186/s40529-017-0161-2. Epub 2017 Jan 9. Bot Stud. 2017. PMID: 28510190 Free PMC article.
-
Novel Alleles of Two Tightly Linked Genes Encoding Polygalacturonase-Inhibiting Proteins (VrPGIP1 and VrPGIP2) Associated with the Br Locus That Confer Bruchid (Callosobruchus spp.) Resistance to Mungbean (Vigna radiata) Accession V2709.Front Plant Sci. 2017 Sep 28;8:1692. doi: 10.3389/fpls.2017.01692. eCollection 2017. Front Plant Sci. 2017. PMID: 29033965 Free PMC article.
-
Physiological, biochemical and molecular evaluation of mungbean genotypes for agronomical yield under drought and salinity stresses in the presence of humic acid.Saudi J Biol Sci. 2022 Sep;29(9):103385. doi: 10.1016/j.sjbs.2022.103385. Epub 2022 Jul 25. Saudi J Biol Sci. 2022. PMID: 35942166 Free PMC article.
-
A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: bruchidae) resistance in mungbean (Vigna radiata).Theor Appl Genet. 2016 Sep;129(9):1673-83. doi: 10.1007/s00122-016-2731-1. Epub 2016 May 24. Theor Appl Genet. 2016. PMID: 27220975
-
Mechanism of Resistance in Mungbean [Vigna radiata (L.) R. Wilczek var. radiata] to bruchids, Callosobruchus spp. (Coleoptera: Bruchidae).Front Plant Sci. 2017 Jun 20;8:1031. doi: 10.3389/fpls.2017.01031. eCollection 2017. Front Plant Sci. 2017. PMID: 28676807 Free PMC article. Review.
References
-
- Bains K, Yang RY, Shanmugasundaram S. High iron mungbean recipes for north India. AVRDC-The World Vegetable Center AVRDC publication, Tainan. 2003;03–562.34:3.
-
- Somta P, Ammaranan C, Ooi PAC, Srinives P. Inheritance of seed resistance to bruchids in cultivated mungbean (Vigna radiata, L. Wilczek) Euphytica. 2007;155(1–2):47–55. doi: 10.1007/s10681-006-9299-9. - DOI
-
- Lambrides CJ, Godwin ID. Mungbean. In: Kole C, editor. Genome mapping and molecular breeding in plants. Pulses, sugar and tuber crops. Germany: Springer; 2007. pp. 69–90.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

