Range of CD4-Bound Conformations of HIV-1 gp120, as Defined Using Conditional CD4-Induced Antibodies
- PMID: 26889042
- PMCID: PMC4836356
- DOI: 10.1128/JVI.03206-15
Range of CD4-Bound Conformations of HIV-1 gp120, as Defined Using Conditional CD4-Induced Antibodies
Abstract
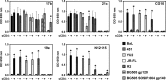
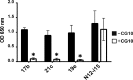
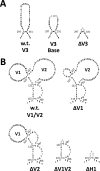
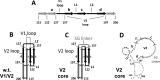
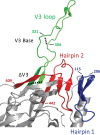
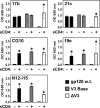
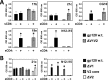
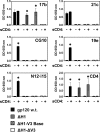
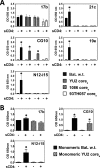
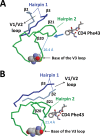
The HIV envelope binds cellular CD4 and undergoes a range of conformational changes that lead to membrane fusion and delivery of the viral nucleocapsid into the cellular cytoplasm. This binding to CD4 reveals cryptic and highly conserved epitopes, the molecular nature of which is still not fully understood. The atomic structures of CD4 complexed with gp120 core molecules (a form of gp120 in which the V1, V2, and V3 loops and N and C termini have been truncated) have indicated that a hallmark feature of the CD4-bound conformation is the bridging sheet minidomain. Variations in the orientation of the bridging sheet hairpins have been revealed when CD4-liganded gp120 was compared to CD4-unliganded trimeric envelope structures. Hence, there appears to be a number of conformational transitions possible in HIV-1 monomeric gp120 that are affected by CD4 binding. The spectrum of CD4-bound conformations has been interrogated in this study by using a well-characterized panel of conditional, CD4-induced (CD4i) monoclonal antibodies (MAbs) that bind HIV-1 gp120 and its mutations under various conditions. Two distinct CD4i epitopes of the outer domain were studied: the first comprises the bridging sheet, while the second contains elements of the V2 loop. Furthermore, we show that the unliganded extended monomeric core of gp120 (coree) assumes an intermediate CD4i conformation in solution that further undergoes detectable rearrangements upon association with CD4. These discoveries impact both accepted paradigms concerning gp120 structure and the field of HIV immunogen design.
Importance: Elucidation of the conformational transitions that the HIV-1 envelope protein undergoes during the course of entry into CD4(+)cells is fundamental to our understanding of HIV biology. The binding of CD4 triggers a range of gp120 structural rearrangements that could present targets for future drug design and development of preventive vaccines. Here we have systematically interrogated and scrutinized these conformational transitions using a panel of antibody probes that share a specific preference for the CD4i conformations. These have been employed to study a collection of gp120 mutations and truncations. Through these analyses, we propose 4 distinct sequential steps in CD4i transitions of gp120 conformations, each defined by antibody specificities and structural requirements of the HIV envelope monomer. As a result, we not only provide new insights into this dynamic process but also define probes to further investigate HIV infection.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- McDougal JS, Mawle A, Cort SP, Nicholson JK, Cross GD, Scheppler-Campbell JA, Hicks D, Sligh J. 1985. Cellular tropism of the human retrovirus HTLV-III/LAV. I. Role of T cell activation and expression of the T4 antigen. J Immunol 135:3151–3162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

