What Makes a Bacterial Species Pathogenic?:Comparative Genomic Analysis of the Genus Leptospira
- PMID: 26890609
- PMCID: PMC4758666
- DOI: 10.1371/journal.pntd.0004403
What Makes a Bacterial Species Pathogenic?:Comparative Genomic Analysis of the Genus Leptospira
Abstract
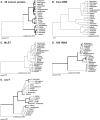
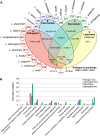
Leptospirosis, caused by spirochetes of the genus Leptospira, is a globally widespread, neglected and emerging zoonotic disease. While whole genome analysis of individual pathogenic, intermediately pathogenic and saprophytic Leptospira species has been reported, comprehensive cross-species genomic comparison of all known species of infectious and non-infectious Leptospira, with the goal of identifying genes related to pathogenesis and mammalian host adaptation, remains a key gap in the field. Infectious Leptospira, comprised of pathogenic and intermediately pathogenic Leptospira, evolutionarily diverged from non-infectious, saprophytic Leptospira, as demonstrated by the following computational biology analyses: 1) the definitive taxonomy and evolutionary relatedness among all known Leptospira species; 2) genomically-predicted metabolic reconstructions that indicate novel adaptation of infectious Leptospira to mammals, including sialic acid biosynthesis, pathogen-specific porphyrin metabolism and the first-time demonstration of cobalamin (B12) autotrophy as a bacterial virulence factor; 3) CRISPR/Cas systems demonstrated only to be present in pathogenic Leptospira, suggesting a potential mechanism for this clade's refractoriness to gene targeting; 4) finding Leptospira pathogen-specific specialized protein secretion systems; 5) novel virulence-related genes/gene families such as the Virulence Modifying (VM) (PF07598 paralogs) proteins and pathogen-specific adhesins; 6) discovery of novel, pathogen-specific protein modification and secretion mechanisms including unique lipoprotein signal peptide motifs, Sec-independent twin arginine protein secretion motifs, and the absence of certain canonical signal recognition particle proteins from all Leptospira; and 7) and demonstration of infectious Leptospira-specific signal-responsive gene expression, motility and chemotaxis systems. By identifying large scale changes in infectious (pathogenic and intermediately pathogenic) vs. non-infectious Leptospira, this work provides new insights into the evolution of a genus of bacterial pathogens. This work will be a comprehensive roadmap for understanding leptospirosis pathogenesis. More generally, it provides new insights into mechanisms by which bacterial pathogens adapt to mammalian hosts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
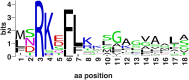




References
-
- Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–71. - PubMed
-
- Ashford DA, Kaiser RM, Spiegel RA, Perkins BA, Weyant RS, Bragg SL, et al. Asymptomatic infection and risk factors for leptospirosis in Nicaragua. Am J Trop Med Hyg. 2000;63(5–6):249–54. . - PubMed
-
- Dikken H, Kmety E. Serological typing methods of leptospires In: B T., NJ R., editors. Methods in Microbiology. 11 London: Academic Press; 1978. p. 259–307.
Publication types
MeSH terms
Substances
Grants and funding
- D43 TW010540/TW/FIC NIH HHS/United States
- D43TW007120/TW/FIC NIH HHS/United States
- R01AI108276/AI/NIAID NIH HHS/United States
- U19AI115658/AI/NIAID NIH HHS/United States
- R21AI115273/AI/NIAID NIH HHS/United States
- K24AI068903/AI/NIAID NIH HHS/United States
- R25TW009338/TW/FIC NIH HHS/United States
- R21 AI115273/AI/NIAID NIH HHS/United States
- R01 AI121207/AI/NIAID NIH HHS/United States
- K24 AI068903/AI/NIAID NIH HHS/United States
- U19 AI115658/AI/NIAID NIH HHS/United States
- U01 AI088752/AI/NIAID NIH HHS/United States
- R01TW009504/TW/FIC NIH HHS/United States
- R01AI121207/AI/NIAID NIH HHS/United States
- R01 AI052473/AI/NIAID NIH HHS/United States
- R25 TW009338/TW/FIC NIH HHS/United States
- HHSN272200900007C/AI/NIAID NIH HHS/United States
- R01AI052473/AI/NIAID NIH HHS/United States
- R01 AI034431/AI/NIAID NIH HHS/United States
- U01AI088752/AI/NIAID NIH HHS/United States
- R01 AI108276/AI/NIAID NIH HHS/United States
- R01 TW009504/TW/FIC NIH HHS/United States
- D43 TW007120/TW/FIC NIH HHS/United States
- I01 BX002003/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

