Transcriptome-Wide Analysis of Hepatitis B Virus-Mediated Changes to Normal Hepatocyte Gene Expression
- PMID: 26891448
- PMCID: PMC4758756
- DOI: 10.1371/journal.ppat.1005438
Transcriptome-Wide Analysis of Hepatitis B Virus-Mediated Changes to Normal Hepatocyte Gene Expression
Abstract
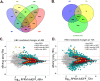
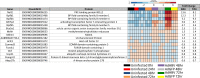
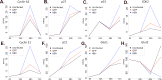
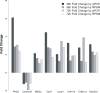
Globally, a chronic hepatitis B virus (HBV) infection remains the leading cause of primary liver cancer. The mechanisms leading to the development of HBV-associated liver cancer remain incompletely understood. In part, this is because studies have been limited by the lack of effective model systems that are both readily available and mimic the cellular environment of a normal hepatocyte. Additionally, many studies have focused on single, specific factors or pathways that may be affected by HBV, without addressing cell physiology as a whole. Here, we apply RNA-seq technology to investigate transcriptome-wide, HBV-mediated changes in gene expression to identify single factors and pathways as well as networks of genes and pathways that are affected in the context of HBV replication. Importantly, these studies were conducted in an ex vivo model of cultured primary hepatocytes, allowing for the transcriptomic characterization of this model system and an investigation of early HBV-mediated effects in a biologically relevant context. We analyzed differential gene expression within the context of time-mediated gene-expression changes and show that in the context of HBV replication a number of genes and cellular pathways are altered, including those associated with metabolism, cell cycle regulation, and lipid biosynthesis. Multiple analysis pipelines, as well as qRT-PCR and an independent, replicate RNA-seq analysis, were used to identify and confirm differentially expressed genes. HBV-mediated alterations to the transcriptome that we identified likely represent early changes to hepatocytes following an HBV infection, suggesting potential targets for early therapeutic intervention. Overall, these studies have produced a valuable resource that can be used to expand our understanding of the complex network of host-virus interactions and the impact of HBV-mediated changes to normal hepatocyte physiology on viral replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22(33):5093–107. Epub 2003/08/12. - PubMed
-
- GLOBOCAN 2012 v1.0 [Internet]. IARC CancerBase No. 11. 2013. Available from: http://globocan.iarc.fr.
-
- Gish RG, Given BD, Lai CL, Locarnini SA, Lau JY, Lewis DL, et al. Chronic hepatitis B: virology, natural history, current management and a glimpse at future opportunities. Antiviral research. 2015. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

