miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9
- PMID: 26893673
- PMCID: PMC4734049
- DOI: 10.3892/ol.2015.4000
miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9
Abstract
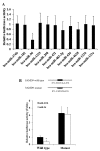
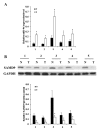
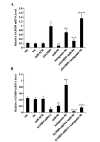
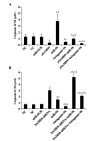
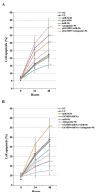
Cisplatin is effective as a single agent or in combination with other drugs for the treatment of non-small cell lung cancer (NSCLC). A concerning clinical challenge with cisplatin-based NSCLC chemotherapy is the intrinsic and acquired chemoresistance to cisplatin. The sterile α motif domain-containing (SAMD9) gene has been reported as a potent tumor suppressor gene that inhibits tumorigenesis and progression of NSCLC. microRNAs (miRNA) have been revealed to play important roles in the regulation of cancer chemoresistance. To the best of our knowledge the present study explored the role of miRNA/SAMD9 signaling in regulating cisplatin chemoresistance in NSCLC for the first time. Out of the several candidate miRNAs predicted to bind the 3'-untranslated region (UTR) of the SAMD9 gene, miRNA-96 (miR-96) demonstrated significant target-sequence-specific inhibition of the SAMD9 3'-UTR luciferase reporter activity in NSCLC cells. In addition, while NSCLC tumor samples exhibited significantly higher expression levels of miR-96 compared with adjacent normal tissues, the expression levels of SAMD9 were significantly lower than those in adjacent normal tissues. miR-96 and SAMD9 were overexpressed and knocked down in the human NSCLC H358 and H23 cell lines and the half maximal inhibitory concentration (IC50) of cisplatin and cell apoptosis rate under cisplatin treatment were used as measures of cisplatin chemoresistance. The present results identified that overexpression of miR-96 in NSCLC cells markedly decreased SAMD9 expression and cisplatin-induced apoptosis, and increased the cisplatin IC50, which could be eliminated by overexpression of SAMD9. By contrast, knocking down miR-96 in NSCLC cells using antagomir-96 significantly increased SAMD9 expression and the cisplatin-induced apoptosis and decreased cisplatin IC50, which could be completely reversed by a knockdown of SAMD9. In conclusion, the current study demonstrates that miR-96 targets and downregulates SAMD9 in NSCLC, which decreases cisplatin-induced apoptosis and induces cisplatin chemoresistance in NSCLC cells. The findings of the present study add novel insights into the function of miR-96 and SAMD9 in cancer, as well as into the molecular mechanisms underlying NSCLC chemoresistance.
Keywords: SAMD9; apoptosis; chemoresistance; cisplatin; miR-96; non-small cell lung cancer.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
