Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. family GH3 β-D-glucosidases
- PMID: 26894673
- PMCID: PMC4756609
- DOI: 10.1107/S2059798315024237
Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. family GH3 β-D-glucosidases
Abstract
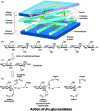
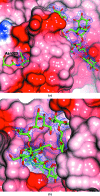
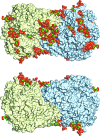
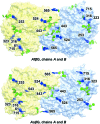
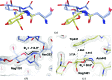
The industrial conversion of cellulosic plant biomass into useful products such as biofuels is a major societal goal. These technologies harness diverse plant degrading enzymes, classical exo- and endo-acting cellulases and, increasingly, cellulose-active lytic polysaccharide monooxygenases, to deconstruct the recalcitrant β-D-linked polysaccharide. A major drawback with this process is that the exo-acting cellobiohydrolases suffer from severe inhibition from their cellobiose product. β-D-Glucosidases are therefore important for liberating glucose from cellobiose and thereby relieving limiting product inhibition. Here, the three-dimensional structures of two industrially important family GH3 β-D-glucosidases from Aspergillus fumigatus and A. oryzae, solved by molecular replacement and refined at 1.95 Å resolution, are reported. Both enzymes, which share 78% sequence identity, display a three-domain structure with the catalytic domain at the interface, as originally shown for barley β-D-glucan exohydrolase, the first three-dimensional structure solved from glycoside hydrolase family GH3. Both enzymes show extensive N-glycosylation, with only a few external sites being truncated to a single GlcNAc molecule. Those glycans N-linked to the core of the structure are identified purely as high-mannose trees, and establish multiple hydrogen bonds between their sugar components and adjacent protein side chains. The extensive glycans pose special problems for crystallographic refinement, and new techniques and protocols were developed especially for this work. These protocols ensured that all of the D-pyranosides in the glycosylation trees were modelled in the preferred minimum-energy (4)C1 chair conformation and should be of general application to refinements of other crystal structures containing O- or N-glycosylation. The Aspergillus GH3 structures, in light of other recent three-dimensional structures, provide insight into fungal β-D-glucosidases and provide a platform on which to inform and inspire new generations of variant enzymes for industrial application.
Keywords: N-glycan; biofuels; cellulose degradation; glucosidase; glycoblocks.
Figures


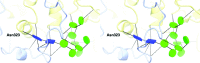
Similar articles
-
Crystal structures of glycoside hydrolase family 3 β-glucosidase 1 from Aspergillus aculeatus.Biochem J. 2013 Jun 1;452(2):211-21. doi: 10.1042/BJ20130054. Biochem J. 2013. PMID: 23537284
-
Crystal Structure of a GH3 β-Glucosidase from the Thermophilic Fungus Chaetomium thermophilum.Int J Mol Sci. 2019 Nov 27;20(23):5962. doi: 10.3390/ijms20235962. Int J Mol Sci. 2019. PMID: 31783503 Free PMC article.
-
Deciphering domain structures of Aspergillus and Streptomyces GH3-β-Glucosidases: a screening system for enzyme engineering and biotechnological applications.BMC Res Notes. 2024 Sep 10;17(1):257. doi: 10.1186/s13104-024-06896-4. BMC Res Notes. 2024. PMID: 39256846 Free PMC article.
-
Dissecting the catalytic mechanism of a plant beta-D-glucan glucohydrolase through structural biology using inhibitors and substrate analogues.Carbohydr Res. 2007 Sep 3;342(12-13):1613-23. doi: 10.1016/j.carres.2007.05.013. Epub 2007 May 18. Carbohydr Res. 2007. PMID: 17548065 Review.
-
Genomics review of holocellulose deconstruction by aspergilli.Microbiol Mol Biol Rev. 2014 Dec;78(4):588-613. doi: 10.1128/MMBR.00019-14. Microbiol Mol Biol Rev. 2014. PMID: 25428936 Free PMC article. Review.
Cited by
-
The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.Nat Commun. 2022 Sep 23;13(1):5577. doi: 10.1038/s41467-022-33180-5. Nat Commun. 2022. PMID: 36151080 Free PMC article.
-
Calcitonin Receptor N-Glycosylation Enhances Peptide Hormone Affinity by Controlling Receptor Dynamics.J Mol Biol. 2020 Mar 27;432(7):1996-2014. doi: 10.1016/j.jmb.2020.01.028. Epub 2020 Feb 6. J Mol Biol. 2020. PMID: 32035902 Free PMC article.
-
Discovery of novel carbohydrate degrading enzymes from soda lakes through functional metagenomics.Front Microbiol. 2022 Dec 7;13:1059061. doi: 10.3389/fmicb.2022.1059061. eCollection 2022. Front Microbiol. 2022. PMID: 36569080 Free PMC article.
-
The enzymatic characters of heterologous expressed novel β-1, 4-glucosidase originated from Aspergillus fresenii.3 Biotech. 2020 Jun;10(6):239. doi: 10.1007/s13205-020-02229-x. Epub 2020 May 8. 3 Biotech. 2020. PMID: 32405443 Free PMC article.
-
Powerful cell wall biomass degradation enzymatic system from saprotrophic Aspergillus fumigatus.Cell Surf. 2024 May 21;11:100126. doi: 10.1016/j.tcsw.2024.100126. eCollection 2024 Jun. Cell Surf. 2024. PMID: 38827922 Free PMC article. Review.
References
-
- Agirre, J., Davies, G., Wilson, K. & Cowtan, K. (2015). Nature Chem. Biol. 11, 303. - PubMed
-
- Agirre, J., Iglesias-Fernandez, J., Rovira, C., Davies, G., Wilson, K. & Cowtan, K. (2015). Nature Struct. Mol. Biol. 22, 833–834. - PubMed
-
- Agrawal, R., Satlewal, A., Gaur, R., Mathur, A., Kumar, R., Gupta, R. P. & Tuli, D. K. (2015). Biochem. Eng. J. 102, 54–61.
-
- Ask, M., Olofsson, K., Di Felice, T., Ruohonen, L., Penttilä, M., Lidén, G. & Olsson, L. (2012). Process Biochem. 47, 1452–1459.
-
- Bacik, J.-P., Whitworth, G. E., Stubbs, K. A., Vocadlo, D. J. & Mark, B. L. (2012). Chem. Biol. 19, 1471–1482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

