The Development of a Recombinant scFv Monoclonal Antibody Targeting Canine CD20 for Use in Comparative Medicine
- PMID: 26894679
- PMCID: PMC4760772
- DOI: 10.1371/journal.pone.0148366
The Development of a Recombinant scFv Monoclonal Antibody Targeting Canine CD20 for Use in Comparative Medicine
Abstract
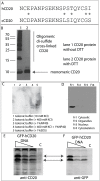
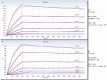
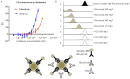
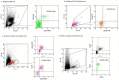
Monoclonal antibodies are leading agents for therapeutic treatment of human diseases, but are limited in use by the paucity of clinically relevant models for validation. Sporadic canine tumours mimic the features of some human equivalents. Developing canine immunotherapeutics can be an approach for modeling human disease responses. Rituximab is a pioneering agent used to treat human hematological malignancies. Biologic mimics that target canine CD20 are just being developed by the biotechnology industry. Towards a comparative canine-human model system, we have developed a novel anti-CD20 monoclonal antibody (NCD1.2) that binds both human and canine CD20. NCD1.2 has a sub-nanomolar Kd as defined by an octet red binding assay. Using FACS, NCD1.2 binds to clinically derived canine cells including B-cells in peripheral blood and in different histotypes of B-cell lymphoma. Immunohistochemical staining of canine tissues indicates that the NCD1.2 binds to membrane localized cells in Diffuse Large B-cell lymphoma, Marginal Zone Lymphoma, and other canine B-cell lymphomas. We cloned the heavy and light chains of NCD1.2 from hybridomas to determine whether active scaffolds can be acquired as future biologics tools. The VH and VL genes from the hybridomas were cloned using degenerate primers and packaged as single chains (scFv) into a phage-display library. Surprisingly, we identified two scFv (scFv-3 and scFv-7) isolated from the hybridoma with bioactivity towards CD20. The two scFv had identical VH genes but different VL genes and identical CDR3s, indicating that at least two light chain mRNAs are encoded by NCD1.2 hybridoma cells. Both scFv-3 and scFv-7 were cloned into mammalian vectors for secretion in CHO cells and the antibodies were bioactive towards recombinant CD20 protein or peptide. The scFv-3 and scFv-7 were cloned into an ADEPT-CPG2 bioconjugate vector where bioactivity was retained when expressed in bacterial systems. These data identify a recombinant anti-CD20 scFv that might form a useful tool for evaluation in bioconjugate-directed anti-CD20 immunotherapies in comparative medicine.
Conflict of interest statement
Figures


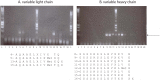
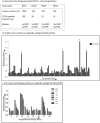
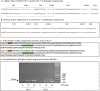

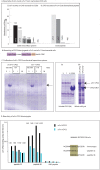
References
-
- Osburn B, Scott C, Gibbs P (2009) One world—one medicine—one health: emerging veterinary challenges and opportunities. Revue scientifique et technique 28: 481–486. - PubMed
-
- Torgerson PR (2013) One world health: socioeconomic burden and parasitic disease control priorities. Veterinary parasitology 195: 223–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

