Systems Level Analyses Reveal Multiple Regulatory Activities of CodY Controlling Metabolism, Motility and Virulence in Listeria monocytogenes
- PMID: 26895237
- PMCID: PMC4760761
- DOI: 10.1371/journal.pgen.1005870
Systems Level Analyses Reveal Multiple Regulatory Activities of CodY Controlling Metabolism, Motility and Virulence in Listeria monocytogenes
Abstract
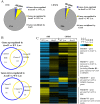
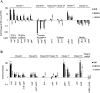
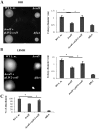
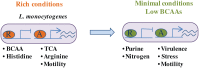
Bacteria sense and respond to many environmental cues, rewiring their regulatory network to facilitate adaptation to new conditions/niches. Global transcription factors that co-regulate multiple pathways simultaneously are essential to this regulatory rewiring. CodY is one such global regulator, controlling expression of both metabolic and virulence genes in Gram-positive bacteria. Branch chained amino acids (BCAAs) serve as a ligand for CodY and modulate its activity. Classically, CodY was considered to function primarily as a repressor under rich growth conditions. However, our previous studies of the bacterial pathogen Listeria monocytogenes revealed that CodY is active also when the bacteria are starved for BCAAs. Under these conditions, CodY loses the ability to repress genes (e.g., metabolic genes) and functions as a direct activator of the master virulence regulator gene, prfA. This observation raised the possibility that CodY possesses multiple functions that allow it to coordinate gene expression across a wide spectrum of metabolic growth conditions, and thus better adapt bacteria to the mammalian niche. To gain a deeper understanding of CodY's regulatory repertoire and identify direct target genes, we performed a genome wide analysis of the CodY regulon and DNA binding under both rich and minimal growth conditions, using RNA-Seq and ChIP-Seq techniques. We demonstrate here that CodY is indeed active (i.e., binds DNA) under both conditions, serving as a repressor and activator of different genes. Further, we identified new genes and pathways that are directly regulated by CodY (e.g., sigB, arg, his, actA, glpF, gadG, gdhA, poxB, glnR and fla genes), integrating metabolism, stress responses, motility and virulence in L. monocytogenes. This study establishes CodY as a multifaceted factor regulating L. monocytogenes physiology in a highly versatile manner.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA.Mol Microbiol. 2015 Feb;95(4):624-44. doi: 10.1111/mmi.12890. Epub 2014 Dec 30. Mol Microbiol. 2015. PMID: 25430920 Free PMC article.
-
CodY-Mediated c-di-GMP-Dependent Inhibition of Mammalian Cell Invasion in Listeria monocytogenes.J Bacteriol. 2018 Feb 7;200(5):e00457-17. doi: 10.1128/JB.00457-17. Print 2018 Mar 1. J Bacteriol. 2018. PMID: 29229701 Free PMC article.
-
Role of GlnR in Controlling Expression of Nitrogen Metabolism Genes in Listeria monocytogenes.J Bacteriol. 2020 Sep 8;202(19):e00209-20. doi: 10.1128/JB.00209-20. Print 2020 Sep 8. J Bacteriol. 2020. PMID: 32690554 Free PMC article.
-
The CodY pleiotropic repressor controls virulence in gram-positive pathogens.FEMS Immunol Med Microbiol. 2011 Jul;62(2):123-39. doi: 10.1111/j.1574-695X.2011.00812.x. Epub 2011 May 27. FEMS Immunol Med Microbiol. 2011. PMID: 21539625 Review.
-
Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria.Int J Med Microbiol. 2014 Mar;304(2):150-5. doi: 10.1016/j.ijmm.2013.11.013. Epub 2013 Dec 1. Int J Med Microbiol. 2014. PMID: 24462007 Review.
Cited by
-
Thermal and Nutritional Regulation of Ribosome Hibernation in Staphylococcus aureus.J Bacteriol. 2018 Nov 26;200(24):e00426-18. doi: 10.1128/JB.00426-18. Print 2018 Dec 15. J Bacteriol. 2018. PMID: 30297357 Free PMC article.
-
A computational system for identifying operons based on RNA-seq data.Methods. 2020 Apr 1;176:62-70. doi: 10.1016/j.ymeth.2019.03.026. Epub 2019 Apr 4. Methods. 2020. PMID: 30953757 Free PMC article. Review.
-
Pseudomonas savastanoi Two-Component System RhpRS Switches between Virulence and Metabolism by Tuning Phosphorylation State and Sensing Nutritional Conditions.mBio. 2019 Mar 19;10(2):e02838-18. doi: 10.1128/mBio.02838-18. mBio. 2019. PMID: 30890603 Free PMC article.
-
Impact of growth pH and glucose concentrations on the CodY regulatory network in Streptococcus salivarius.BMC Genomics. 2018 May 23;19(1):386. doi: 10.1186/s12864-018-4781-z. BMC Genomics. 2018. PMID: 29792173 Free PMC article.
-
Regulation of Listeria monocytogenes Virulence.Microbiol Spectr. 2019 Jul;7(4):10.1128/microbiolspec.gpp3-0064-2019. doi: 10.1128/microbiolspec.GPP3-0064-2019. Microbiol Spectr. 2019. PMID: 31441398 Free PMC article. Review.
References
-
- Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65(7):1127–41. - PubMed
-
- Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Molecular microbiology. 1995;16(2):251–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

