Releasing Activity Disengages Cohesin's Smc3/Scc1 Interface in a Process Blocked by Acetylation
- PMID: 26895425
- PMCID: PMC4769318
- DOI: 10.1016/j.molcel.2016.01.026
Releasing Activity Disengages Cohesin's Smc3/Scc1 Interface in a Process Blocked by Acetylation
Abstract
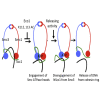
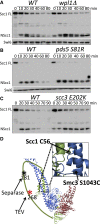
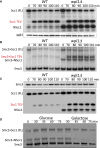
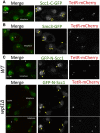
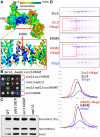
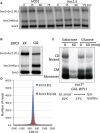
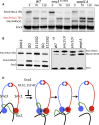
Sister chromatid cohesion conferred by entrapment of sister DNAs within a tripartite ring formed between cohesin's Scc1, Smc1, and Smc3 subunits is created during S and destroyed at anaphase through Scc1 cleavage by separase. Cohesin's association with chromosomes is controlled by opposing activities: loading by Scc2/4 complex and release by a separase-independent releasing activity as well as by cleavage. Coentrapment of sister DNAs at replication is accompanied by acetylation of Smc3 by Eco1, which blocks releasing activity and ensures that sisters remain connected. Because fusion of Smc3 to Scc1 prevents release and bypasses the requirement for Eco1, we suggested that release is mediated by disengagement of the Smc3/Scc1 interface. We show that mutations capable of bypassing Eco1 in Smc1, Smc3, Scc1, Wapl, Pds5, and Scc3 subunits reduce dissociation of N-terminal cleavage fragments of Scc1 (NScc1) from Smc3. This process involves interaction between Smc ATPase heads and is inhibited by Smc3 acetylation.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Arumugam P., Gruber S., Tanaka K., Haering C.H., Mechtler K., Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003;13:1941–1953. - PubMed
-
- Arumugam P., Nishino T., Haering C.H., Gruber S., Nasmyth K. Cohesin’s ATPase activity is stimulated by the C-terminal Winged-Helix domain of its kleisin subunit. Curr. Biol. 2006;16:1998–2008. - PubMed
-
- Bürmann F., Shin H.C., Basquin J., Soh Y.M., Giménez-Oya V., Kim Y.G., Oh B.H., Gruber S. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat. Struct. Mol. Biol. 2013;20:371–379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

