Regulator of G Protein Signaling 7 (RGS7) Can Exist in a Homo-oligomeric Form That Is Regulated by Gαo and R7-binding Protein
- PMID: 26895961
- PMCID: PMC4861480
- DOI: 10.1074/jbc.M115.694075
Regulator of G Protein Signaling 7 (RGS7) Can Exist in a Homo-oligomeric Form That Is Regulated by Gαo and R7-binding Protein
Abstract
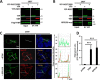
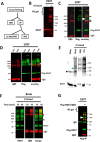
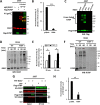
RGS (regulator of G protein signaling) proteins of the R7 subfamily (RGS6, -7, -9, and -11) are highly expressed in neurons where they regulate many physiological processes. R7 RGS proteins contain several distinct domains and form obligatory dimers with the atypical Gβ subunit, Gβ5 They also interact with other proteins such as R7-binding protein, R9-anchoring protein, and the orphan receptors GPR158 and GPR179. These interactions facilitate plasma membrane targeting and stability of R7 proteins and modulate their activity. Here, we investigated RGS7 complexes using in situ chemical cross-linking. We found that in mouse brain and transfected cells cross-linking causes formation of distinct RGS7 complexes. One of the products had the apparent molecular mass of ∼150 kDa on SDS-PAGE and did not contain Gβ5 Mass spectrometry analysis showed no other proteins to be present within the 150-kDa complex in the amount close to stoichiometric with RGS7. This finding suggested that RGS7 could form a homo-oligomer. Indeed, co-immunoprecipitation of differentially tagged RGS7 constructs, with or without chemical cross-linking, demonstrated RGS7 self-association. RGS7-RGS7 interaction required the DEP domain but not the RGS and DHEX domains or the Gβ5 subunit. Using transfected cells and knock-out mice, we demonstrated that R7-binding protein had a strong inhibitory effect on homo-oligomerization of RGS7. In contrast, our data indicated that GPR158 could bind to the RGS7 homo-oligomer without causing its dissociation. Co-expression of constitutively active Gαo prevented the RGS7-RGS7 interaction. These results reveal the existence of RGS protein homo-oligomers and show regulation of their assembly by R7 RGS-binding partners.
Keywords: G protein; G protein-coupled receptor (GPCR); R7-binding protein (R7BP); immunoprecipitation; oligomer; protein cross-linking; regulator of G protein signaling (RGS); subcellular localization.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
Regulation of neurite morphogenesis by interaction between R7 regulator of G protein signaling complexes and G protein subunit Gα13.J Biol Chem. 2017 Jun 16;292(24):9906-9918. doi: 10.1074/jbc.M116.771923. Epub 2017 Apr 21. J Biol Chem. 2017. PMID: 28432124 Free PMC article.
-
Macromolecular composition dictates receptor and G protein selectivity of regulator of G protein signaling (RGS) 7 and 9-2 protein complexes in living cells.J Biol Chem. 2013 Aug 30;288(35):25129-25142. doi: 10.1074/jbc.M113.462283. Epub 2013 Jul 15. J Biol Chem. 2013. PMID: 23857581 Free PMC article.
-
Subcellular localization of regulator of G protein signaling RGS7 complex in neurons and transfected cells.J Neurochem. 2012 Aug;122(3):568-81. doi: 10.1111/j.1471-4159.2012.07811.x. Epub 2012 Jun 22. J Neurochem. 2012. PMID: 22640015 Free PMC article.
-
Structure, function, and localization of Gβ5-RGS complexes.Prog Mol Biol Transl Sci. 2009;86:157-203. doi: 10.1016/S1877-1173(09)86006-7. Epub 2009 Oct 7. Prog Mol Biol Transl Sci. 2009. PMID: 20374716 Free PMC article. Review.
-
The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling.Cell Biochem Biophys. 2009;54(1-3):33-46. doi: 10.1007/s12013-009-9052-9. Epub 2009 Jun 12. Cell Biochem Biophys. 2009. PMID: 19521673 Free PMC article. Review.
Cited by
-
Identification of DEP domain-containing proteins by a machine learning method and experimental analysis of their expression in human HCC tissues.Sci Rep. 2016 Dec 21;6:39655. doi: 10.1038/srep39655. Sci Rep. 2016. PMID: 28000796 Free PMC article.
-
Regulation of neurite morphogenesis by interaction between R7 regulator of G protein signaling complexes and G protein subunit Gα13.J Biol Chem. 2017 Jun 16;292(24):9906-9918. doi: 10.1074/jbc.M116.771923. Epub 2017 Apr 21. J Biol Chem. 2017. PMID: 28432124 Free PMC article.
-
Osteocalcin and GPR158: linking bone and brain function.Front Cell Dev Biol. 2025 Apr 23;13:1564751. doi: 10.3389/fcell.2025.1564751. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40337551 Free PMC article. Review.
-
Non-Ceruloplasmin Copper Identifies a Subtype of Alzheimer's Disease (CuAD): Characterization of the Cognitive Profile and Case of a CuAD Patient Carrying an RGS7 Stop-Loss Variant.Int J Mol Sci. 2023 Mar 28;24(7):6377. doi: 10.3390/ijms24076377. Int J Mol Sci. 2023. PMID: 37047347 Free PMC article.
-
Expression Mapping and Functional Analysis of Orphan G-Protein-Coupled Receptor GPR158 in the Adult Mouse Brain Using a GPR158 Transgenic Mouse.Biomolecules. 2023 Mar 5;13(3):479. doi: 10.3390/biom13030479. Biomolecules. 2023. PMID: 36979415 Free PMC article.
References
-
- Neer E. J. (1995) Heterotrimeric G proteins: organizers of transmembrane signals. Cell 80, 249–257 - PubMed
-
- Cabrera-Vera T. M., Vanhauwe J., Thomas T. O., Medkova M., Preininger A., Mazzoni M. R., and Hamm H. E. (2003) Insights into G protein structure, function, and regulation. Endocr. Rev. 24, 765–781 - PubMed
-
- Clapham D. E., and Neer E. J. (1997) G protein βγ subunits. Annu. Rev. Pharmacol. Toxicol. 37, 167–203 - PubMed
-
- Lan K. L., Zhong H., Nanamori M., and Neubig R. R. (2000) Rapid kinetics of regulator of G-protein signaling (RGS)-mediated Gαi and Gαo deactivation. Gα specificity of RGS4 and RGS7. J. Biol. Chem. 275, 33497–33503 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

