Engineering Escherichia coli for Microbial Production of Butanone
- PMID: 26896132
- PMCID: PMC4836414
- DOI: 10.1128/AEM.03964-15
Engineering Escherichia coli for Microbial Production of Butanone
Abstract
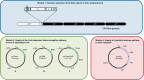
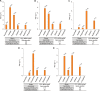
To expand the chemical and molecular diversity of biotransformation using whole-cell biocatalysts, we genetically engineered a pathway in Escherichia coli for heterologous production of butanone, an important commodity ketone. First, a 1-propanol-producing E. coli host strain with its sleeping beauty mutase (Sbm) operon being activated was used to increase the pool of propionyl-coenzyme A (propionyl-CoA). Subsequently, molecular heterofusion of propionyl-CoA and acetyl-CoA was conducted to yield 3-ketovaleryl-CoA via a CoA-dependent elongation pathway. Lastly, 3-ketovaleryl-CoA was channeled into the clostridial acetone formation pathway for thioester hydrolysis and subsequent decarboxylation to form butanone. Biochemical, genetic, and metabolic factors affecting relative levels of ketogenesis, acidogenesis, and alcohol genesis under selected fermentative culture conditions were investigated. Using the engineered E. coli strain for batch cultivation with 30 g liter(-1)glycerol as the carbon source, we achieved coproduction of 1.3 g liter(-1)butanone and 2.9 g liter(-1)acetone. The results suggest that approximately 42% of spent glycerol was utilized for ketone biosynthesis, and thus they demonstrate potential industrial applicability of this microbial platform.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Engineering of Escherichia coli for direct and modulated biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer using unrelated carbon sources.Sci Rep. 2016 Nov 7;6:36470. doi: 10.1038/srep36470. Sci Rep. 2016. PMID: 27819347 Free PMC article.
-
Biochemical, genetic, and metabolic engineering strategies to enhance coproduction of 1-propanol and ethanol in engineered Escherichia coli.Appl Microbiol Biotechnol. 2014 Nov;98(22):9499-515. doi: 10.1007/s00253-014-6093-9. Epub 2014 Oct 10. Appl Microbiol Biotechnol. 2014. PMID: 25301579
-
Integrated strain engineering and bioprocessing strategies for high-level bio-based production of 3-hydroxyvalerate in Escherichia coli.Appl Microbiol Biotechnol. 2020 Jun;104(12):5259-5272. doi: 10.1007/s00253-020-10580-5. Epub 2020 Apr 14. Appl Microbiol Biotechnol. 2020. PMID: 32291486
-
Engineering Escherichia coli Cell Factories for n-Butanol Production.Adv Biochem Eng Biotechnol. 2016;155:141-63. doi: 10.1007/10_2015_306. Adv Biochem Eng Biotechnol. 2016. PMID: 25662903 Review.
-
Engineering metabolic pathways in Escherichia coli for constructing a "microbial chassis" for biochemical production.Bioresour Technol. 2017 Dec;245(Pt B):1362-1368. doi: 10.1016/j.biortech.2017.05.008. Epub 2017 May 4. Bioresour Technol. 2017. PMID: 28522199 Review.
Cited by
-
High-throughput colorimetric assays optimized for detection of ketones and aldehydes produced by microbial cell factories.Microb Biotechnol. 2022 Sep;15(9):2426-2438. doi: 10.1111/1751-7915.14097. Epub 2022 Jun 10. Microb Biotechnol. 2022. PMID: 35689383 Free PMC article.
-
Metabolic engineering of β-oxidation to leverage thioesterases for production of 2-heptanone, 2-nonanone and 2-undecanone.Metab Eng. 2020 Sep;61:335-343. doi: 10.1016/j.ymben.2020.05.008. Epub 2020 May 29. Metab Eng. 2020. PMID: 32479802 Free PMC article.
-
Feasible-metabolic-pathway-exploration technique using chemical latent space.Bioinformatics. 2020 Dec 30;36(Suppl_2):i770-i778. doi: 10.1093/bioinformatics/btaa809. Bioinformatics. 2020. PMID: 33381845 Free PMC article.
-
Short-chain ketone production by engineered polyketide synthases in Streptomyces albus.Nat Commun. 2018 Nov 1;9(1):4569. doi: 10.1038/s41467-018-07040-0. Nat Commun. 2018. PMID: 30385744 Free PMC article.
-
Synergy at work: linking the metabolism of two lactic acid bacteria to achieve superior production of 2-butanol.Biotechnol Biofuels. 2020 Mar 11;13:45. doi: 10.1186/s13068-020-01689-w. eCollection 2020. Biotechnol Biofuels. 2020. PMID: 32180827 Free PMC article.
References
-
- Hoell D, Mensing T, Roggenbuck R, Sakuth M, Sperlich E, Urban T, Neier W, Strehlke G. 2000. 2-Butanone. Wiley-VCH Verlag, Weinheim, Germany.
-
- Ghiaci P, Norbeck J, Larsson C. 2014. 2-Butanol and butanone production in Saccharomyces cerevisiae through combination of a B12 dependent dehydratase and a secondary alcohol dehydrogenase using a TEV-based expression system. PLoS One 9:e102774. doi:10.1371/journal.pone.0102774. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

