Molecular foundations of chilling-tolerance of modern maize
- PMID: 26897027
- PMCID: PMC4761173
- DOI: 10.1186/s12864-016-2453-4
Molecular foundations of chilling-tolerance of modern maize
Abstract
Background: Recent progress in selective breeding of maize (Zea mays L.) towards adaptation to temperate climate has allowed the production of inbred lines withstanding cold springs with temperatures below 8 °C or even close to 0 °C, indicating that despite its tropical origins maize is not inherently cold-sensitive.
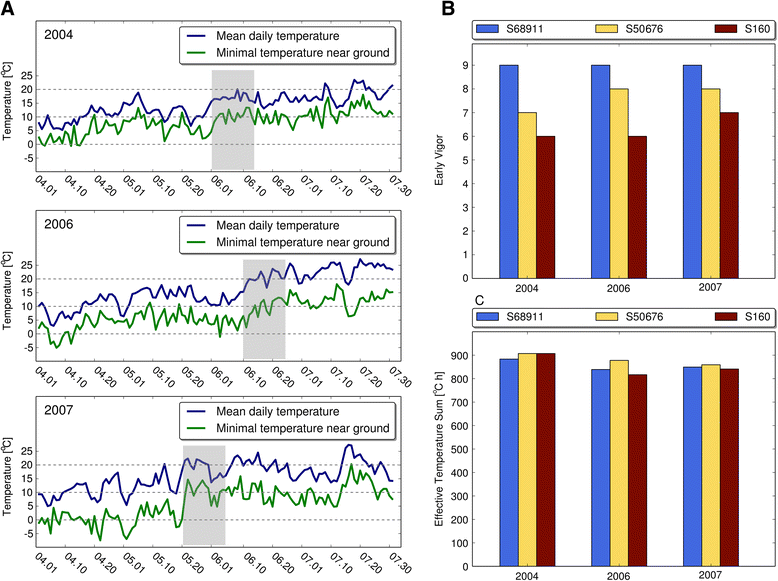
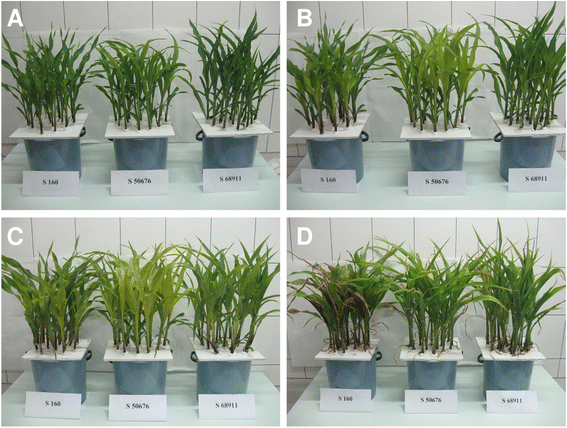
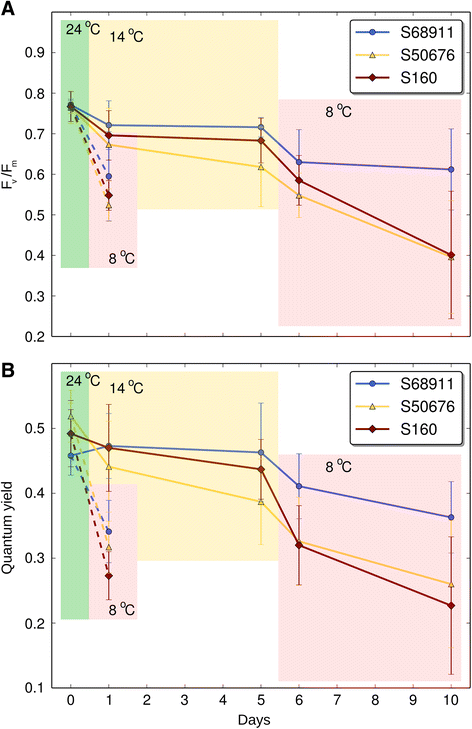
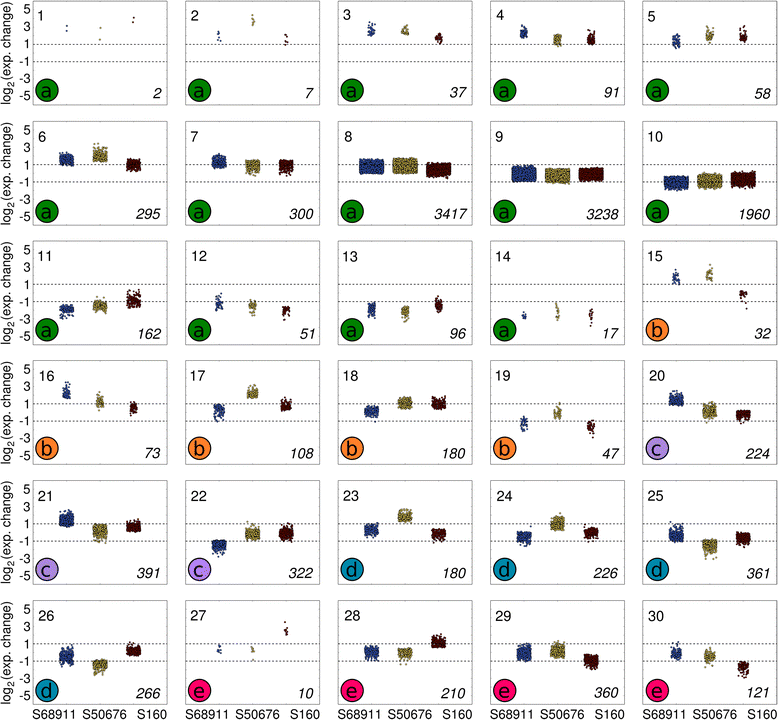
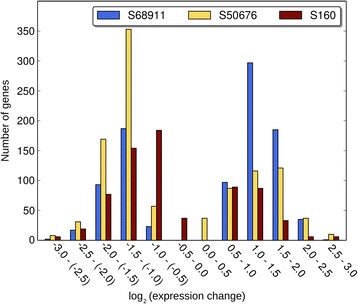
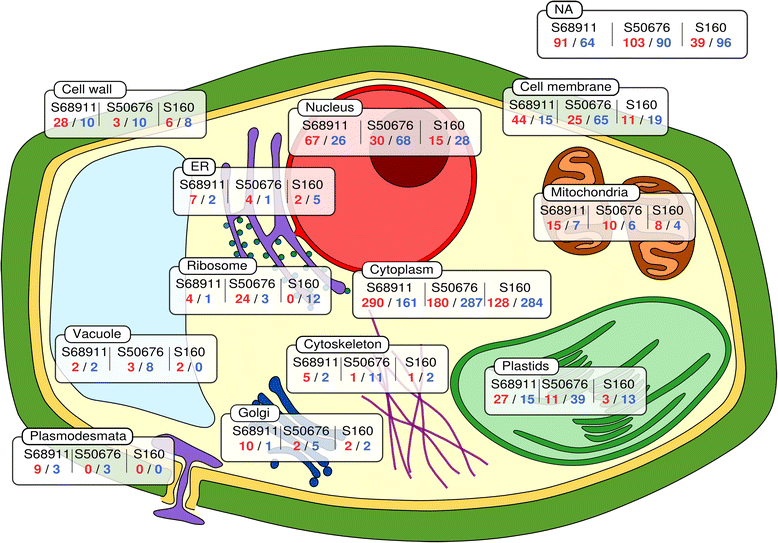
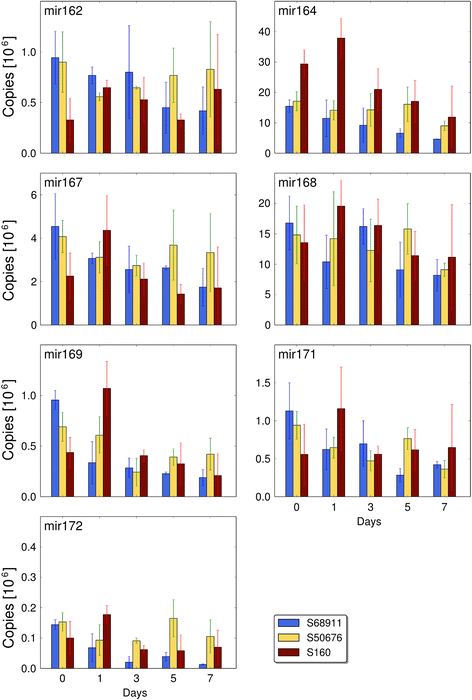
Results: Here we studied the acclimatory response of three maize inbred lines of contrasting cold-sensitivity selected basing on multi-year routine field data. The field observations were confirmed in the growth chamber. Under controlled conditions the damage to the photosynthetic apparatus due to severe cold treatment was the least in the cold-tolerant line provided that it had been subjected to prior moderate chilling, i.e., acclimation. The cold-sensitive lines performed equally poorly with or without acclimation. To uncover the molecular basis of the attained cold-acclimatability we performed comparative transcriptome profiling of the response of the lines to the cold during acclimation phase by means of microarrays with a statistical and bioinformatic data analysis.
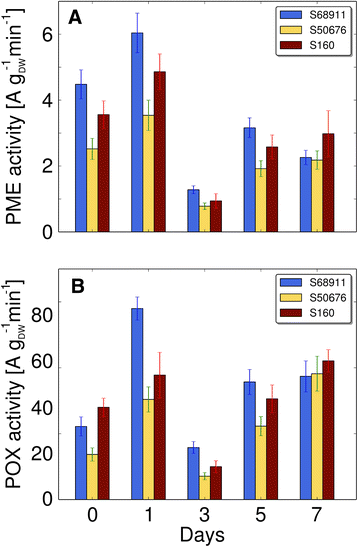
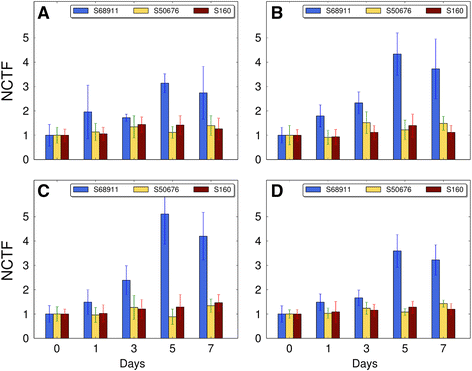
Conclusions: The analyses indicated three mechanisms likely responsible for the cold-tolerance: acclimation-dependent modification of the photosynthetic apparatus, cell wall properties, and developmental processes. Those conclusions supported the observed acclimation of photosynthesis to severe cold at moderate chilling and were further confirmed by experimentally showing specific modification of cell wall properties and repression of selected miRNA species, general regulators of development, in the cold-tolerant line subjected to cold stress.
Figures
References
-
- Troyer AF. Breeding adapted, popular maize hybrids. In: Tigersteld PMA, editor. Adaptation in Plant Breeding. Netherlands: Kluwer Academic Publishers; 1997. pp. 185–96.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

