Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity
- PMID: 26898111
- PMCID: PMC4871114
- DOI: 10.1038/nri.2016.2
Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity
Abstract
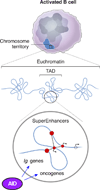
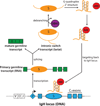
As B cells engage in the immune response, they express activation-induced cytidine deaminase (AID) to initiate the hypermutation and recombination of immunoglobulin genes, which are crucial processes for the efficient recognition and disposal of pathogens. However, AID must be tightly controlled in B cells to minimize off-target mutations, which can drive chromosomal translocations and the development of B cell malignancies, such as lymphomas. Recent genomic and biochemical analyses have begun to unravel the mechanisms of how AID-mediated deamination is targeted outside immunoglobulin genes. Here, we discuss the transcriptional and topological features that are emerging as key drivers of AID promiscuous activity.
Figures



References
-
- Pieper K, Grimbacher B, Eibel H. B-cell biology and development. J Allergy Clin Immunol. 2013;131:959–971. - PubMed
-
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. - PubMed
-
- Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2)[see comments] Cell. 2000;102:565–575. - PubMed
-
- Muramatsu M, et al. Class switch recombination hypermutation require activation-induced cytidine deaminase (AID), a potential RNAediting enzyme [see comments] Cell. 2000;102:553–563. - PubMed
-
- Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

