DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution
- PMID: 26899170
- PMCID: PMC4762164
- DOI: 10.1186/s13059-016-0893-4
DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution
Abstract
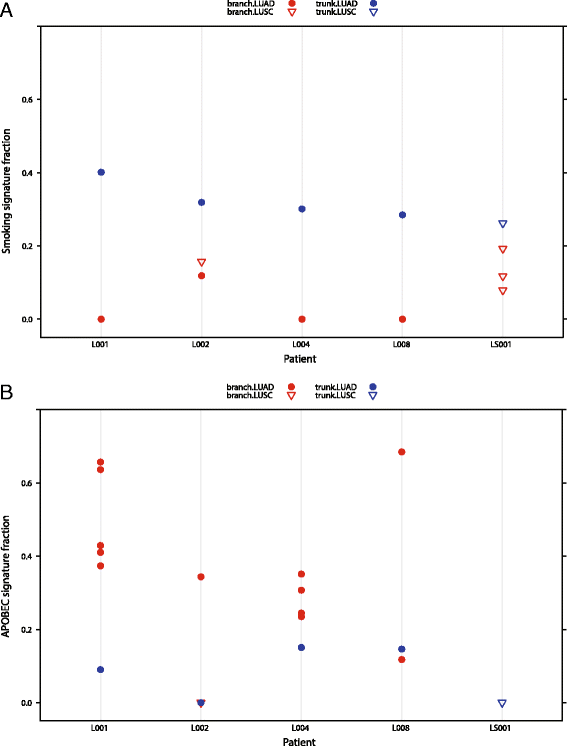
Background: Analysis of somatic mutations provides insight into the mutational processes that have shaped the cancer genome, but such analysis currently requires large cohorts. We develop deconstructSigs, which allows the identification of mutational signatures within a single tumor sample.
Results: Application of deconstructSigs identifies samples with DNA repair deficiencies and reveals distinct and dynamic mutational processes molding the cancer genome in esophageal adenocarcinoma compared to squamous cell carcinomas.
Conclusions: deconstructSigs confers the ability to define mutational processes driven by environmental exposures, DNA repair abnormalities, and mutagenic processes in individual tumors with implications for precision cancer medicine.
Figures




References
-
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013 - PubMed
-
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell. 2013. http://dx.doi.org/10.1016/j.celrep.2012.12.008. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

