Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle
- PMID: 26900483
- PMCID: PMC4759782
- DOI: 10.1186/s40851-016-0039-2
Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle
Abstract
Introduction: Bivalve molluscs have flourished in marine environments, and many species constitute important aquatic resources. Recently, whole genome sequences from two bivalves, the pearl oyster, Pinctada fucata, and the Pacific oyster, Crassostrea gigas, have been decoded, making it possible to compare genomic sequences among molluscs, and to explore general and lineage-specific genetic features and trends in bivalves. In order to improve the quality of sequence data for these purposes, we have updated the entire P. fucata genome assembly.
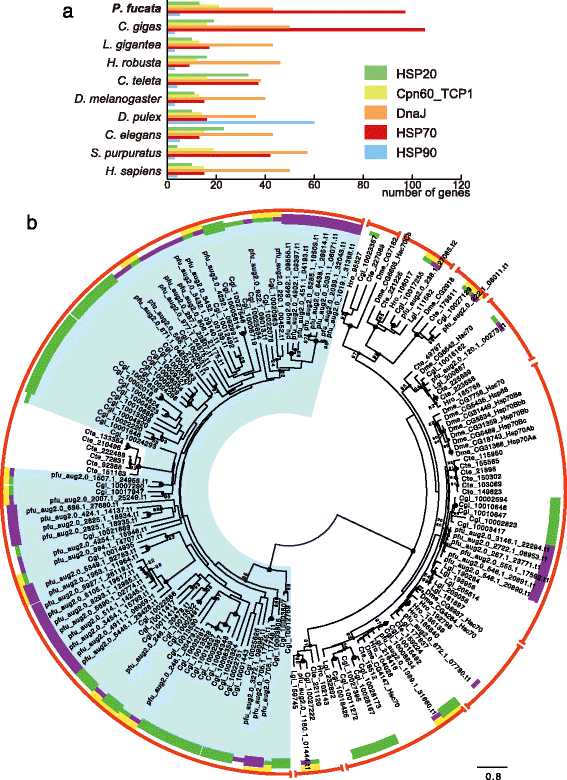
Results: We present a new genome assembly of the pearl oyster, Pinctada fucata (version 2.0). To update the assembly, we conducted additional sequencing, obtaining accumulated sequence data amounting to 193× the P. fucata genome. Sequence redundancy in contigs that was caused by heterozygosity was removed in silico, which significantly improved subsequent scaffolding. Gene model version 2.0 was generated with the aid of manual gene annotations supplied by the P. fucata research community. Comparison of mollusc and other bilaterian genomes shows that gene arrangements of Hox, ParaHox, and Wnt clusters in the P. fucata genome are similar to those of other molluscs. Like the Pacific oyster, P. fucata possesses many genes involved in environmental responses and in immune defense. Phylogenetic analyses of heat shock protein70 and C1q domain-containing protein families indicate that extensive expansion of genes occurred independently in each lineage. Several gene duplication events prior to the split between the pearl oyster and the Pacific oyster are also evident. In addition, a number of tandem duplications of genes that encode shell matrix proteins are also well characterized in the P. fucata genome.
Conclusions: Both the Pinctada and Crassostrea lineages have expanded specific gene families in a lineage-specific manner. Frequent duplication of genes responsible for shell formation in the P. fucata genome explains the diversity of mollusc shell structures. These duplications reveal dynamic genome evolution to forge the complex physiology that enables bivalves to employ a sessile lifestyle in the intertidal zone.
Keywords: Biomineralization; C1q; Genome; Heat shock proteins; Hox; ParaHox; Pearl oyster; Pinctada fucata.
Figures





References
-
- Bayne BL, Newell RC. Physiological energetics of marine molluscs. In: Wilbur KM, Saleuddin ASM, editors. The Mollusca. 4. Cambridge: Academic; 1983. pp. 407–515.
-
- Gerdol M, Manfrin C, De Moro G, Figueras A, Novoa B, Venier P, et al. The C1q domain containing proteins of the Mediterranean mussel Mytilus galloprovincialis: A widespread and diverse family of immune-related molecules. Dev Comp Immunol. 2011;35(6):635–43. doi: 10.1016/j.dci.2011.01.018. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

