Identification of Differentially Expressed Genes Related to Dehydration Resistance in a Highly Drought-Tolerant Pear, Pyrus betulaefolia, as through RNA-Seq
- PMID: 26900681
- PMCID: PMC4762547
- DOI: 10.1371/journal.pone.0149352
Identification of Differentially Expressed Genes Related to Dehydration Resistance in a Highly Drought-Tolerant Pear, Pyrus betulaefolia, as through RNA-Seq
Retraction in
-
Retraction: Identification of Differentially Expressed Genes Related to Dehydration Resistance in a Highly Drought-Tolerant Pear, Pyrus betulaefolia, as through RNA-Seq.PLoS One. 2024 May 22;19(5):e0304430. doi: 10.1371/journal.pone.0304430. eCollection 2024. PLoS One. 2024. PMID: 38776316 Free PMC article. No abstract available.
Abstract
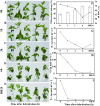
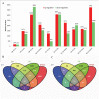
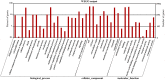
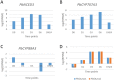
Drought is a major abiotic stress that affects plant growth, development and productivity. Pear is one of the most important deciduous fruit trees in the world, but the mechanisms of drought tolerance in this plant are still unclear. To better understand the molecular basis regarding drought stress response, RNA-seq was performed on samples collected before and after dehydration in Pyrus betulaefolia. In total, 19,532 differentially expressed genes (DEGs) were identified. These genes were annotated into 144 Gene Ontology (GO) terms and 18 clusters of orthologous groups (COG) involved in 129 Kyoto Encyclopedia of Genes and Genomes (KEGG) defined pathways. These DEGs comprised 49 (26 up-regulated, 23 down-regulated), 248 (166 up-regulated, 82 down-regulated), 3483 (1295 up-regulated, 2188 down-regulated), 1455 (1065 up-regulated, 390 down-regulated) genes from the 1 h, 3 h and 6 h dehydration-treated samples and a 24 h recovery samples, respectively. RNA-seq was validated by analyzing the expresson patterns of randomly selected 16 DEGs by quantitative real-time PCR. Photosynthesis, signal transduction, innate immune response, protein phosphorylation, response to water, response to biotic stimulus, and plant hormone signal transduction were the most significantly enriched GO categories amongst the DEGs. A total of 637 transcription factors were shown to be dehydration responsive. In addition, a number of genes involved in the metabolism and signaling of hormones were significantly affected by the dehydration stress. This dataset provides valuable information regarding the Pyrus betulaefolia transcriptome changes in response to dehydration and may promote identification and functional analysis of potential genes that could be used for improving drought tolerance via genetic engineering of non-model, but economically-important, perennial species.
Conflict of interest statement
Figures

References
-
- Aswath CR, Kim SH, Mo SY, Kim DH. Transgenic plants of creeping bent grass harboring the stress inducible gene, 9-cis-epoxycarotenoid dioxygenase, are highly tolerant to drought and NaCl stress. Plant growth regulation. 2005;47(2–3):129–39. 10.1007/s10725-005-3380-6 - DOI
-
- Bailly C, Audigier C, Ladonne F, Wagner MH, Coste F, Corbineau F, et al. Changes in oligosaccharide content and antioxidant enzyme activities in developing bean seeds as related to acquisition of drying tolerance and seed quality. Journal of experimental botany. 2001;52(357):701–8. . - PubMed
-
- Wang YC, Jiang J, Zhao X, Liu GF, Yang CP, Zhan LP. A novel LEA gene from Tamarix androssowii confers drought tolerance in transgenic tobacco. Plant Science. 2006;171(6):655–62. 10.1016/j.plantsci.2006.06.011 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

