Lattice engineering through nanoparticle-DNA frameworks
- PMID: 26901516
- PMCID: PMC5282967
- DOI: 10.1038/nmat4571
Lattice engineering through nanoparticle-DNA frameworks
Abstract
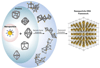
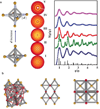
Advances in self-assembly over the past decade have demonstrated that nano- and microscale particles can be organized into a large diversity of ordered three-dimensional (3D) lattices. However, the ability to generate different desired lattice types from the same set of particles remains challenging. Here, we show that nanoparticles can be assembled into crystalline and open 3D frameworks by connecting them through designed DNA-based polyhedral frames. The geometrical shapes of the frames, combined with the DNA-assisted binding properties of their vertices, facilitate the well-defined topological connections between particles in accordance with frame geometry. With this strategy, different crystallographic lattices using the same particles can be assembled by introduction of the corresponding DNA polyhedral frames. This approach should facilitate the rational assembly of nanoscale lattices through the design of the unit cell.
Figures


References
-
- Talapin DV, Lee J-S, Kovalenko MV, Shevchenko EV. Prospects of Colloidal Nanocrystals for Electronic and Optoelectronic Applications. Chem. Rev. 2010;110:389–458. - PubMed
-
- Hu T, Isaacoff BP, Bahng JH, Hao C, Zhou Y, Zhu J, Li X, Wang Z, Liu S, Xu C, Biteen JS, Kotov NA. Self-Organization of Plasmonic and Excitonic Nanoparticles into Resonant Chiral Supraparticle Assemblies. Nano Lett. 2014;14:6799–6810. - PubMed
-
- Cargnello M, Johnston-Peck AC, Diroll BT, Wong E, Datta B, Damodhar D, Doan-Nguyen VVT, Herzing AA, Kagan CR, Murray CB. Substitutional doping in nanocrystal superlattices. Nature. 2015;524:450–453. - PubMed
-
- Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller E-M, Högele A, Simmel FC, Govorov AO, Liedl T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012;483:311–314. - PubMed
-
- Xiong H, Sfeir MY, Gang O. Assembly, structure and optical response of three-dimensional dynamically tunable multicomponent superlattices. Nano Lett. 2010;10:4456–4462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

