Lymphatic endothelial cells are a replicative niche for Mycobacterium tuberculosis
- PMID: 26901813
- PMCID: PMC4767353
- DOI: 10.1172/JCI83379
Lymphatic endothelial cells are a replicative niche for Mycobacterium tuberculosis
Abstract
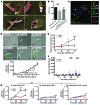
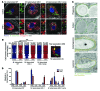
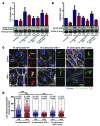
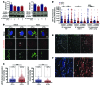
In extrapulmonary tuberculosis, the most common site of infection is within the lymphatic system, and there is growing recognition that lymphatic endothelial cells (LECs) are involved in immune function. Here, we identified LECs, which line the lymphatic vessels, as a niche for Mycobacterium tuberculosis in the lymph nodes of patients with tuberculosis. In cultured primary human LECs (hLECs), we determined that M. tuberculosis replicates both in the cytosol and within autophagosomes, but the bacteria failed to replicate when the virulence locus RD1 was deleted. Activation by IFN-γ induced a cell-autonomous response in hLECs via autophagy and NO production that restricted M. tuberculosis growth. Thus, depending on the activation status of LECs, autophagy can both promote and restrict replication. Together, these findings reveal a previously unrecognized role for hLECs and autophagy in tuberculosis pathogenesis and suggest that hLECs are a potential niche for M. tuberculosis that allows establishment of persistent infection in lymph nodes.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

