Population structure and acquisition of the vanB resistance determinant in German clinical isolates of Enterococcus faecium ST192
- PMID: 26902259
- PMCID: PMC4763178
- DOI: 10.1038/srep21847
Population structure and acquisition of the vanB resistance determinant in German clinical isolates of Enterococcus faecium ST192
Abstract
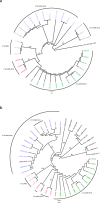
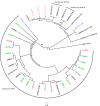
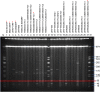
In the context of the global action plan to reduce the dissemination of antibiotic resistances it is of utmost importance to understand the population structure of resistant endemic bacterial lineages and to elucidate how bacteria acquire certain resistance determinants. Vancomycin resistant enterococci represent one such example of a prominent nosocomial pathogen on which nation-wide population analyses on prevalent lineages are scarce and data on how the bacteria acquire resistance, especially of the vanB genotype, are still under debate. With respect to Germany, an increased prevalence of VRE was noted in recent years. Here, invasive infections caused by sequence type ST192 VRE are often associated with the vanB-type resistance determinant. Hence, we analyzed 49 vanB-positive and vanB-negative E. faecium isolates by means of whole genome sequencing. Our studies revealed a distinct population structure and that spread of the Tn1549-vanB-type resistance involves exchange of large chromosomal fragments between vanB-positive and vanB-negative enterococci rather than independent acquisition events. In vitro filter-mating experiments support the hypothesis and suggest the presence of certain target sequences as a limiting factor for dissemination of the vanB element. Thus, the present study provides a better understanding of how enterococci emerge into successful multidrug-resistant nosocomial pathogens.
Figures

References
-
- Centers for Disease, C. & Prevention. Nosocomial enterococci resistant to vancomycin–United States, 1989–1993. MMWR Morb Mortal Wkly Rep 42, 597–599 (1993). - PubMed
-
- Schouten M. A., Hoogkamp-Korstanje J. A., Meis J. F. & Voss A. & European, V. R. E. S. G. Prevalence of vancomycin-resistant enterococci in Europe. Eur J Clin Microbiol Infect Dis 19, 816–822 (2000). - PubMed
-
- Garnier F., Taourit S., Glaser P., Courvalin P. & Galimand M. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146 (Pt 6), 1481–1489 (2000). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

