Transcriptional identification and characterization of differentially expressed genes associated with embryogenesis in radish (Raphanus sativus L.)
- PMID: 26902837
- PMCID: PMC4763228
- DOI: 10.1038/srep21652
Transcriptional identification and characterization of differentially expressed genes associated with embryogenesis in radish (Raphanus sativus L.)
Abstract
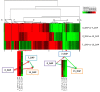
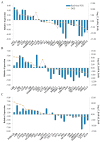
Embryogenesis is an important component in the life cycle of most plant species. Due to the difficulty in embryo isolation, the global gene expression involved in plant embryogenesis, especially the early events following fertilization are largely unknown in radish. In this study, three cDNA libraries from ovules of radish before and after fertilization were sequenced using the Digital Gene Expression (DGE) tag profiling strategy. A total of 5,777 differentially expressed transcripts were detected based on pairwise comparison in the three libraries (0_DAP, 7_DAP and 15_DAP). Results from Gene Ontology (GO) and pathway enrichment analysis revealed that these differentially expressed genes (DEGs) were implicated in numerous life processes including embryo development and phytohormones biosynthesis. Notably, some genes encoding auxin response factor (ARF ), Leafy cotyledon1 (LEC1) and somatic embryogenesis receptor-like kinase (SERK ) known to be involved in radish embryogenesis were differentially expressed. The expression patterns of 30 genes including LEC1-2, AGL9, LRR, PKL and ARF8-1 were validated by qRT-PCR. Furthermore, the cooperation between miRNA and mRNA may play a pivotal role in the radish embryogenesis process. This is the first report on identification of DEGs profiles related to radish embryogenesis and seed development. These results could facilitate further dissection of the molecular mechanisms underlying embryogenesis and seed development in radish.
Figures





References
-
- Radoeva T. & Weijers D. A roadmap to embryo identity in plants. Trends Plant Sci. 19, 709–716 (2014). - PubMed
-
- Smertenko A. & Bozhkov P. V. Somatic embryogenesis: life and death processes during apical-basal patterning. J. Exp. Bot. 65, 1343–1360 (2014). - PubMed
-
- ten Hove C. A., Lu K. J. & Weijers D. Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142, 420–430 (2015). - PubMed
-
- Goldberg R. B., Paiva G. d. & Yadegari R. Plant embryogenesis: zygote to seed science. Science 266, 605–614 (1994). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

