T cell-specific inactivation of mouse CD2 by CRISPR/Cas9
- PMID: 26903281
- PMCID: PMC4763270
- DOI: 10.1038/srep21377
T cell-specific inactivation of mouse CD2 by CRISPR/Cas9
Abstract
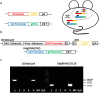
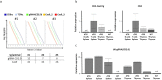
The CRISPR/Cas9 system can be used to mutate target sequences by introduction of double-strand breaks followed by imprecise repair. To test its use for conditional gene editing we generated mice transgenic for CD4 promoter-driven Cas9 combined with guide RNA targeting CD2. We found that within CD4(+) and CD8(+) lymphocytes from lymph nodes and spleen 1% and 0.6% were not expressing CD2, respectively. T cells lacking CD2 carryied mutations, which confirmed that Cas9 driven by cell-type specific promoters can edit genes in the mouse and may thus allow targeted studies of gene function in vivo.
Figures




References
-
- Hooper M., Hardy K., Handyside A., Hunter S. & Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326, 292 (1987). - PubMed
-
- Gu H., Marth J. D., Orban P. C., Mossmann H. & Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting [see comments]. Science 265, 103–106 (1994). - PubMed
-
- Kühn R., Schwenk F., Aguet M. & Rajewsky K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

