Structural Features of Ion Transport and Allosteric Regulation in Sodium-Calcium Exchanger (NCX) Proteins
- PMID: 26903880
- PMCID: PMC4746289
- DOI: 10.3389/fphys.2016.00030
Structural Features of Ion Transport and Allosteric Regulation in Sodium-Calcium Exchanger (NCX) Proteins
Abstract
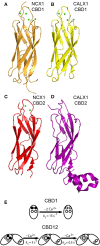
Na(+)/Ca(2+) exchanger (NCX) proteins extrude Ca(2+) from the cell to maintain cellular homeostasis. Since NCX proteins contribute to numerous physiological and pathophysiological events, their pharmacological targeting has been desired for a long time. This intervention remains challenging owing to our poor understanding of the underlying structure-dynamic mechanisms. Recent structural studies have shed light on the structure-function relationships underlying the ion-transport and allosteric regulation of NCX. The crystal structure of an archaeal NCX (NCX_Mj) along with molecular dynamics simulations and ion flux analyses, have assigned the ion binding sites for 3Na(+) and 1Ca(2+), which are being transported in separate steps. In contrast with NCX_Mj, eukaryotic NCXs contain the regulatory Ca(2+)-binding domains, CBD1 and CBD2, which affect the membrane embedded ion-transport domains over a distance of ~80 Å. The Ca(2+)-dependent regulation is ortholog, isoform, and splice-variant dependent to meet physiological requirements, exhibiting either a positive, negative, or no response to regulatory Ca(2+). The crystal structures of the two-domain (CBD12) tandem have revealed a common mechanism involving a Ca(2+)-driven tethering of CBDs in diverse NCX variants. However, dissociation kinetics of occluded Ca(2+) (entrapped at the two-domain interface) depends on the alternative-splicing segment (at CBD2), thereby representing splicing-dependent dynamic coupling of CBDs. The HDX-MS, SAXS, NMR, FRET, equilibrium (45)Ca(2+) binding and stopped-flow techniques provided insights into the dynamic mechanisms of CBDs. Ca(2+) binding to CBD1 results in a population shift, where more constraint conformational states become highly populated without global conformational changes in the alignment of CBDs. This mechanism is common among NCXs. Recent HDX-MS studies have demonstrated that the apo CBD1 and CBD2 are stabilized by interacting with each other, while Ca(2+) binding to CBD1 rigidifies local backbone segments of CBD2, but not of CBD1. The extent and strength of Ca(2+)-dependent rigidification at CBD2 is splice-variant dependent, showing clear correlations with phenotypes of matching NCX variants. Therefore, diverse NCX variants share a common mechanism for the initial decoding of the regulatory signal upon Ca(2+) binding at the interface of CBDs, whereas the allosteric message is shaped by CBD2, the dynamic features of which are dictated by the splicing segment.
Keywords: Ca2+ binding proteins; HDX-MS; NCX; SAXS; X-ray crystallography; allosteric regulation.
Figures




References
-
- Besserer G. M., Ottolia M., Nicoll D. A., Chaptal V., Cascio D., Philipson K. D., et al. . (2007). The second Ca2+-binding domain of the Na+ Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis. Proc. Natl. Acad. Sci. U.S.A. 104, 18467–18472. 10.1073/pnas.0707417104 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

