Hydrogenase Gene Distribution and H2 Consumption Ability within the Thiomicrospira Lineage
- PMID: 26903978
- PMCID: PMC4744846
- DOI: 10.3389/fmicb.2016.00099
Hydrogenase Gene Distribution and H2 Consumption Ability within the Thiomicrospira Lineage
Abstract
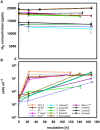
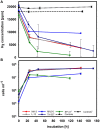
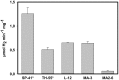
Thiomicrospira were originally characterized as sulfur-oxidizing chemolithoautotrophs. Attempts to grow them on hydrogen failed for many years. Only recently we demonstrated hydrogen consumption among two of three tested Thiomicrospira and posited that hydrogen consumption may be more widespread among Thiomicrospira than previously assumed. Here, we investigate and compare the hydrogen consumption ability and the presence of group 1 [NiFe]-hydrogenase genes (enzyme catalyzes H2↔2H(+) + 2e(-)) for sixteen different Thiomicrospira species. Seven of these Thiomicrospira species encoded group 1 [NiFe]-hydrogenase genes and five of these species could also consume hydrogen. All Thiomicrospira species exhibiting hydrogen consumption were from hydrothermal vents along the Mid-Atlantic ridge or Eastern Pacific ridges. The tested Thiomicrospira from Mediterranean and Western Pacific vents could not consume hydrogen. The [NiFe]-hydrogenase genes were categorized into two clusters: those resembling the hydrogenase from Hydrogenovibrio are in cluster I and are related to those from Alpha- and other Gammaproteobacteria. In cluster II, hydrogenases found exclusively in Thiomicrospira crunogena strains are combined and form a monophyletic group with those from Epsilonproteobacteria suggesting they were acquired through horizontal gene transfer. Hydrogen consumption appears to be common among some Thiomicrospira, given that five of the tested sixteen strains carried this trait. The hydrogen consumption ability expands their competitiveness within an environment.
Keywords: Thiomicrospira; [NiFe]-hydrogenases; biogeography; horizontal gene transfer; hydrogen consumption.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

