PATtyFams: Protein Families for the Microbial Genomes in the PATRIC Database
- PMID: 26903996
- PMCID: PMC4744870
- DOI: 10.3389/fmicb.2016.00118
PATtyFams: Protein Families for the Microbial Genomes in the PATRIC Database
Abstract
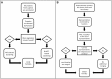
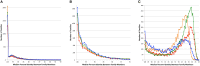
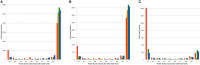
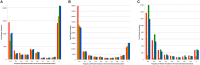
The ability to build accurate protein families is a fundamental operation in bioinformatics that influences comparative analyses, genome annotation, and metabolic modeling. For several years we have been maintaining protein families for all microbial genomes in the PATRIC database (Pathosystems Resource Integration Center, patricbrc.org) in order to drive many of the comparative analysis tools that are available through the PATRIC website. However, due to the burgeoning number of genomes, traditional approaches for generating protein families are becoming prohibitive. In this report, we describe a new approach for generating protein families, which we call PATtyFams. This method uses the k-mer-based function assignments available through RAST (Rapid Annotation using Subsystem Technology) to rapidly guide family formation, and then differentiates the function-based groups into families using a Markov Cluster algorithm (MCL). This new approach for generating protein families is rapid, scalable and has properties that are consistent with alignment-based methods.
Keywords: FIGfams; RAST; comparative genomics; genome annotation; metabolic modeling.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

