Presence of Viral RNA and Proteins in Exosomes from Cellular Clones Resistant to Rift Valley Fever Virus Infection
- PMID: 26904012
- PMCID: PMC4749701
- DOI: 10.3389/fmicb.2016.00139
Presence of Viral RNA and Proteins in Exosomes from Cellular Clones Resistant to Rift Valley Fever Virus Infection
Abstract
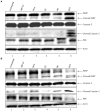
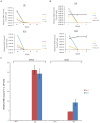
Rift Valley Fever Virus (RVFV) is a RNA virus that belongs to the genus Phlebovirus, family Bunyaviridae. It infects humans and livestock and causes Rift Valley fever. RVFV is considered an agricultural pathogen by the USDA, as it can cause up to 100% abortion in cattle and extensive death of newborns. In addition, it is designated as Category A pathogen by the CDC and the NIAID. In some human cases of RVFV infection, the virus causes fever, ocular damage, liver damage, hemorrhagic fever, and death. There are currently limited options for vaccine candidates, which include the MP-12 and clone 13 versions of RVFV. Viral infections often deregulate multiple cellular pathways that contribute to replication and host pathology. We have previously shown that latent human immunodeficiency virus-1 (HIV-1) and human T-cell lymphotropic virus-1 (HTLV-1) infected cells secrete exosomes that contain short viral RNAs, limited number of genomic RNAs, and viral proteins. These exosomes largely target neighboring cells and activate the NF-κB pathway, leading to cell proliferation, and overall better viral replication. In this manuscript, we studied the effects of exosome formation from RVFV infected cells and their function on recipient cells. We initially infected cells, isolated resistant clones, and further purified using dilution cloning. We then characterized these cells as resistant to new RVFV infection, but sensitive to other viral infections, including Venezuelan Equine Encephalitis Virus (VEEV). These clones contained normal markers (i.e., CD63) for exosomes and were able to activate the TLR pathway in recipient reporter cells. Interestingly, the exosome rich preparations, much like their host cell, contained viral RNA (L, M, and S genome). The RNAs were detected using qRT-PCR in both parental and exosomal preparations as well as in CD63 immunoprecipitates. Viral proteins such as N and a modified form of NSs were present in some of these exosomes. Finally, treatment of recipient cells (T-cells and monocytic cells) showed drastic rate of apoptosis through PARP cleavage and caspase 3 activation from some but not all exosome enriched preparations. Collectively, these data suggest that exosomes from RVFV infected cells alter the dynamics of the immune cells and may contribute to pathology of the viral infection.
Keywords: Rift Valley Fever Virus; exosomes; resistant clones.
Figures





References
-
- Billecocq A., Spiegel M., Vialat P., Kohl A., Weber F., Bouloy M., et al. (2004). NSs protein of rift valley fever virus blocks interferon production by inhibiting host gene transcription NSs protein of rift valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78 9798–9806. 10.1128/JVI.78.18.9798 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

