Computational Prediction of Effector Proteins in Fungi: Opportunities and Challenges
- PMID: 26904083
- PMCID: PMC4751359
- DOI: 10.3389/fpls.2016.00126
Computational Prediction of Effector Proteins in Fungi: Opportunities and Challenges
Abstract
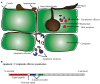
Effector proteins are mostly secretory proteins that stimulate plant infection by manipulating the host response. Identifying fungal effector proteins and understanding their function is of great importance in efforts to curb losses to plant diseases. Recent advances in high-throughput sequencing technologies have facilitated the availability of several fungal genomes and 1000s of transcriptomes. As a result, the growing amount of genomic information has provided great opportunities to identify putative effector proteins in different fungal species. There is little consensus over the annotation and functionality of effector proteins, and mostly small secretory proteins are considered as effector proteins, a concept that tends to overestimate the number of proteins involved in a plant-pathogen interaction. With the characterization of Avr genes, criteria for computational prediction of effector proteins are becoming more efficient. There are 100s of tools available for the identification of conserved motifs, signature sequences and structural features in the proteins. Many pipelines and online servers, which combine several tools, are made available to perform genome-wide identification of effector proteins. In this review, available tools and pipelines, their strength and limitations for effective identification of fungal effector proteins are discussed. We also present an exhaustive list of classically secreted proteins along with their key conserved motifs found in 12 common plant pathogens (11 fungi and one oomycete) through an analytical pipeline.
Keywords: classification and prediction; computational tool and servers; effector proteins; fungal secretome; host–pathogen interaction.
Figures

Similar articles
-
Evaluation of Secretion Prediction Highlights Differing Approaches Needed for Oomycete and Fungal Effectors.Front Plant Sci. 2015 Dec 23;6:1168. doi: 10.3389/fpls.2015.01168. eCollection 2015. Front Plant Sci. 2015. PMID: 26779196 Free PMC article.
-
Using Population and Comparative Genomics to Understand the Genetic Basis of Effector-Driven Fungal Pathogen Evolution.Front Plant Sci. 2017 Feb 3;8:119. doi: 10.3389/fpls.2017.00119. eCollection 2017. Front Plant Sci. 2017. PMID: 28217138 Free PMC article. Review.
-
Effector Biology of Biotrophic Plant Fungal Pathogens: Current Advances and Future Prospects.Microbiol Res. 2020 Dec;241:126567. doi: 10.1016/j.micres.2020.126567. Epub 2020 Aug 23. Microbiol Res. 2020. PMID: 33080488 Review.
-
Effector-Mining in the Poplar Rust Fungus Melampsora larici-populina Secretome.Front Plant Sci. 2015 Dec 15;6:1051. doi: 10.3389/fpls.2015.01051. eCollection 2015. Front Plant Sci. 2015. PMID: 26697026 Free PMC article. Review.
-
Prediction of effector proteins and their implications in pathogenicity of phytopathogenic filamentous fungi: A review.Int J Biol Macromol. 2022 May 1;206:188-202. doi: 10.1016/j.ijbiomac.2022.02.133. Epub 2022 Feb 26. Int J Biol Macromol. 2022. PMID: 35227707 Review.
Cited by
-
Genome-Wide Identification and Characterization of Effector Candidates with Conserved Motif in Falciphora oryzae.Int J Mol Sci. 2024 Jan 4;25(1):650. doi: 10.3390/ijms25010650. Int J Mol Sci. 2024. PMID: 38203820 Free PMC article.
-
Identification of Secretory Proteins in Mycobacterium tuberculosis Using Pseudo Amino Acid Composition.Biomed Res Int. 2016;2016:5413903. doi: 10.1155/2016/5413903. Epub 2016 Aug 11. Biomed Res Int. 2016. PMID: 27597968 Free PMC article.
-
Genome sequencing and annotation of Cercospora sesami, a fungal pathogen causing leaf spot to Sesamum indicum.3 Biotech. 2023 Feb;13(2):55. doi: 10.1007/s13205-023-03468-4. Epub 2023 Jan 19. 3 Biotech. 2023. PMID: 36685323 Free PMC article.
-
Genome-wide analysis of Claviceps paspali: insights into the secretome of the main species causing ergot disease in Paspalum spp.BMC Genomics. 2021 Oct 26;22(1):766. doi: 10.1186/s12864-021-08077-0. BMC Genomics. 2021. PMID: 34702162 Free PMC article.
-
Characterization of Arbuscular Mycorrhizal Effector Proteins.Int J Mol Sci. 2023 May 23;24(11):9125. doi: 10.3390/ijms24119125. Int J Mol Sci. 2023. PMID: 37298075 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

