Constraint Based Modeling Going Multicellular
- PMID: 26904548
- PMCID: PMC4748834
- DOI: 10.3389/fmolb.2016.00003
Constraint Based Modeling Going Multicellular
Abstract
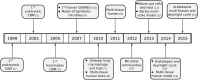
Constraint based modeling has seen applications in many microorganisms. For example, there are now established methods to determine potential genetic modifications and external interventions to increase the efficiency of microbial strains in chemical production pipelines. In addition, multiple models of multicellular organisms have been created including plants and humans. While initially the focus here was on modeling individual cell types of the multicellular organism, this focus recently started to switch. Models of microbial communities, as well as multi-tissue models of higher organisms have been constructed. These models thereby can include different parts of a plant, like root, stem, or different tissue types in the same organ. Such models can elucidate details of the interplay between symbiotic organisms, as well as the concerted efforts of multiple tissues and can be applied to analyse the effects of drugs or mutations on a more systemic level. In this review we give an overview of the recent development of multi-tissue models using constraint based techniques and the methods employed when investigating these models. We further highlight advances in combining constraint based models with dynamic and regulatory information and give an overview of these types of hybrid or multi-level approaches.
Keywords: constraint based modeling; metabolic modeling; multi-organism modeling; multi-scale modeling; multi-tissue modeling.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

