Characterization of FGFR1 Locus in sqNSCLC Reveals a Broad and Heterogeneous Amplicon
- PMID: 26905262
- PMCID: PMC4764357
- DOI: 10.1371/journal.pone.0149628
Characterization of FGFR1 Locus in sqNSCLC Reveals a Broad and Heterogeneous Amplicon
Abstract
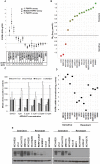
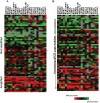
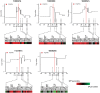
FGFR1 amplification occurs in ~20% of sqNSCLC and trials with FGFR inhibitors have selected FGFR1 amplified patients by FISH. Lung cancer cell lines were profiled for sensitivity to AZD4547, a potent, selective inhibitor of FGFRs 1-3. Sensitivity to FGFR inhibition was associated with but not wholly predicted by increased FGFR1 gene copy number. Additional biomarker assays evaluating expression of FGFRs and correlation between amplification and expression in clinical tissues are therefore warranted. We validated nanoString for mRNA expression analysis of 194 genes, including FGFRs, from clinical tumour tissue. In a panel of sqNSCLC tumours 14.4% (13/90) were FGFR1 amplified by FISH. Although mean FGFR1 expression was significantly higher in amplified samples, there was significant overlap in the range of expression levels between the amplified and non-amplified cohorts with several non-amplified samples expressing FGFR1 to levels equivalent to amplified samples. Statistical analysis revealed increased expression of FGFR1 neighboring genes on the 8p12 amplicon (BAG4, LSM1 and WHSC1L1) in FGFR1 amplified tumours, suggesting a broad rather than focal amplicon and raises the potential for codependencies. High resolution aCGH analysis of pre-clinical and clinical samples supported the presence of a broad and heterogeneous amplicon around the FGFR1 locus. In conclusion, the range of FGFR1 expression levels in both FGFR1 amplified and non-amplified NSCLC tissues, together with the breadth and intra-patient heterogeneity of the 8p amplicon highlights the need for gene expression analysis of clinical samples to inform the understanding of determinants of response to FGFR inhibitors. In this respect the nanoString platform provides an attractive option for RNA analysis of FFPE clinical samples.
Conflict of interest statement
Figures



References
-
- Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. The Lancet Oncology. 2010;11(2):121–8. 10.1016/S1470-2045(09)70364-X . - DOI - PubMed
-
- Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, Engelman JA, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. The Lancet Oncology. 2011;12(11):1004–12. 10.1016/S1470-2045(11)70232-7 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

