Ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
- PMID: 26905735
- PMCID: PMC4765138
- DOI: 10.1186/s40168-016-0153-6
Ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
Abstract
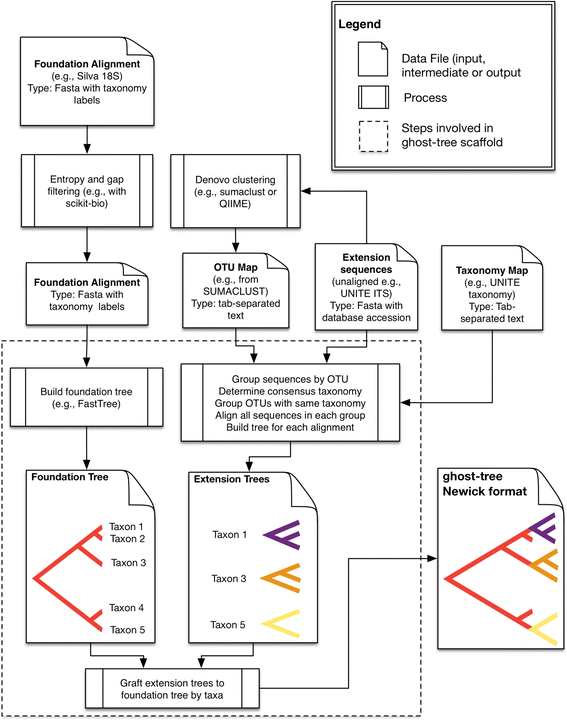
Background: Fungi play critical roles in many ecosystems, cause serious diseases in plants and animals, and pose significant threats to human health and structural integrity problems in built environments. While most fungal diversity remains unknown, the development of PCR primers for the internal transcribed spacer (ITS) combined with next-generation sequencing has substantially improved our ability to profile fungal microbial diversity. Although the high sequence variability in the ITS region facilitates more accurate species identification, it also makes multiple sequence alignment and phylogenetic analysis unreliable across evolutionarily distant fungi because the sequences are hard to align accurately. To address this issue, we created ghost-tree, a bioinformatics tool that integrates sequence data from two genetic markers into a single phylogenetic tree that can be used for diversity analyses. Our approach starts with a "foundation" phylogeny based on one genetic marker whose sequences can be aligned across organisms spanning divergent taxonomic groups (e.g., fungal families). Then, "extension" phylogenies are built for more closely related organisms (e.g., fungal species or strains) using a second more rapidly evolving genetic marker. These smaller phylogenies are then grafted onto the foundation tree by mapping taxonomic names such that each corresponding foundation-tree tip would branch into its new "extension tree" child.
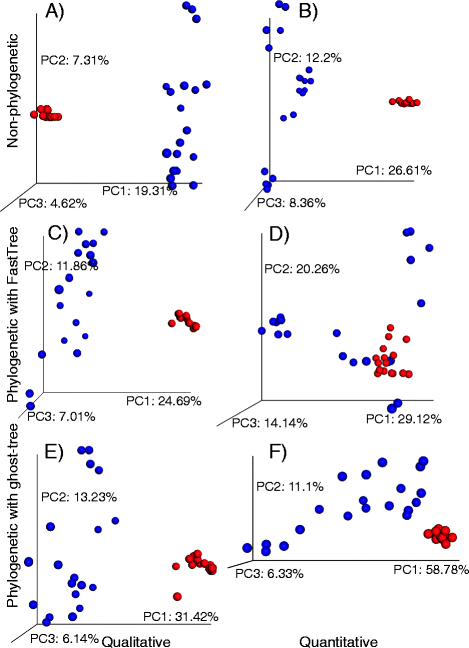
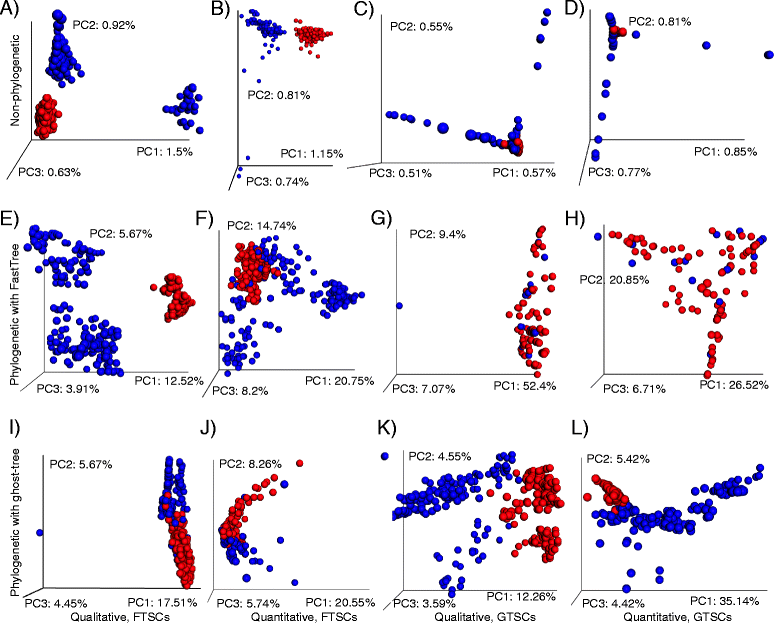
Results: We applied ghost-tree to graft fungal extension phylogenies derived from ITS sequences onto a foundation phylogeny derived from fungal 18S sequences. Our analysis of simulated and real fungal ITS data sets found that phylogenetic distances between fungal communities computed using ghost-tree phylogenies explained significantly more variance than non-phylogenetic distances. The phylogenetic metrics also improved our ability to distinguish small differences (effect sizes) between microbial communities, though results were similar to non-phylogenetic methods for larger effect sizes.
Conclusions: The Silva/UNITE-based ghost tree presented here can be easily integrated into existing fungal analysis pipelines to enhance the resolution of fungal community differences and improve understanding of these communities in built environments. The ghost-tree software package can also be used to develop phylogenetic trees for other marker gene sets that afford different taxonomic resolution, or for bridging genome trees with amplicon trees.
Availability: ghost-tree is pip-installable. All source code, documentation, and test code are available under the BSD license at https://github.com/JTFouquier/ghost-tree .
Figures
Similar articles
-
A phylogenetic framework for the kingdom Fungi based on 18S rRNA gene sequences.Mar Genomics. 2017 Dec;36:33-39. doi: 10.1016/j.margen.2017.05.009. Epub 2017 Jun 1. Mar Genomics. 2017. PMID: 28578827
-
A new promising phylogenetic marker to study the diversity of fungal communities: The Glycoside Hydrolase 63 gene.Mol Ecol Resour. 2017 Nov;17(6):e1-e11. doi: 10.1111/1755-0998.12678. Epub 2017 May 12. Mol Ecol Resour. 2017. PMID: 28382652
-
Utilization of the relative complexity measure to construct a phylogenetic tree for fungi.Mycol Res. 2004 Feb;108(Pt 2):117-25. doi: 10.1017/s0953756203009079. Mycol Res. 2004. PMID: 15119348
-
Fungal DNA barcoding.Genome. 2016 Nov;59(11):913-932. doi: 10.1139/gen-2016-0046. Epub 2016 Aug 30. Genome. 2016. PMID: 27829306 Review.
-
The search for the fungal tree of life.Trends Microbiol. 2009 Nov;17(11):488-97. doi: 10.1016/j.tim.2009.08.001. Epub 2009 Sep 24. Trends Microbiol. 2009. PMID: 19782570 Review.
Cited by
-
A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming.Nat Commun. 2020 Sep 18;11(1):4717. doi: 10.1038/s41467-020-18560-z. Nat Commun. 2020. PMID: 32948774 Free PMC article.
-
Assembly of the Populus Microbiome Is Temporally Dynamic and Determined by Selective and Stochastic Factors.mSphere. 2021 Jun 30;6(3):e0131620. doi: 10.1128/mSphere.01316-20. Epub 2021 Jun 9. mSphere. 2021. PMID: 34106767 Free PMC article.
-
Development of the Human Mycobiome over the First Month of Life and across Body Sites.mSystems. 2018 Mar 6;3(3):e00140-17. doi: 10.1128/mSystems.00140-17. eCollection 2018 May-Jun. mSystems. 2018. PMID: 29546248 Free PMC article.
-
Deciphering Succession and Assembly Patterns of Microbial Communities in a Two-Stage Solid-State Fermentation System.Microbiol Spectr. 2021 Oct 31;9(2):e0071821. doi: 10.1128/Spectrum.00718-21. Epub 2021 Sep 22. Microbiol Spectr. 2021. PMID: 34549993 Free PMC article.
-
Integrating sequence composition information into microbial diversity analyses with k-mer frequency counting.mSystems. 2025 Mar 18;10(3):e0155024. doi: 10.1128/msystems.01550-24. Epub 2025 Feb 20. mSystems. 2025. PMID: 39976436 Free PMC article.
References
-
- van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396(6706):69–72. doi: 10.1038/23932. - DOI
-
- Centers for Disease Control and Prevention . CDC and fungal diseases: why are fungal diseases a public health issue? 2009.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

