Crystal Structure of Alcohol Oxidase from Pichia pastoris
- PMID: 26905908
- PMCID: PMC4764120
- DOI: 10.1371/journal.pone.0149846
Crystal Structure of Alcohol Oxidase from Pichia pastoris
Abstract
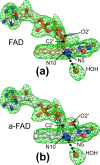
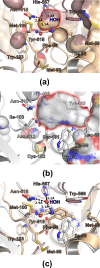
FAD-dependent alcohol oxidases (AOX) are key enzymes of methylotrophic organisms that can utilize lower primary alcohols as sole source of carbon and energy. Here we report the crystal structure analysis of the methanol oxidase AOX1 from Pichia pastoris. The crystallographic phase problem was solved by means of Molecular Replacement in combination with initial structure rebuilding using Rosetta model completion and relaxation against an averaged electron density map. The subunit arrangement of the homo-octameric AOX1 differs from that of octameric vanillyl alcohol oxidase and other dimeric or tetrameric alcohol oxidases, due to the insertion of two large protruding loop regions and an additional C-terminal extension in AOX1. In comparison to other alcohol oxidases, the active site cavity of AOX1 is significantly reduced in size, which could explain the observed preference for methanol as substrate. All AOX1 subunits of the structure reported here harbor a modified flavin adenine dinucleotide, which contains an arabityl chain instead of a ribityl chain attached to the isoalloxazine ring.
Conflict of interest statement
Figures



Similar articles
-
Crystal structure of p-hydroxybenzoate hydroxylase reconstituted with the modified FAD present in alcohol oxidase from methylotrophic yeasts: evidence for an arabinoflavin.Protein Sci. 1994 Dec;3(12):2245-53. doi: 10.1002/pro.5560031210. Protein Sci. 1994. PMID: 7756982 Free PMC article.
-
Structure of Alcohol Oxidase from Pichia pastoris by Cryo-Electron Microscopy.PLoS One. 2016 Jul 26;11(7):e0159476. doi: 10.1371/journal.pone.0159476. eCollection 2016. PLoS One. 2016. PMID: 27458710 Free PMC article.
-
Formation and distribution of modified FAD between isozymes of alcohol oxidase in the methylotrophic yeast pichia methanolica.Biochemistry (Mosc). 1998 Dec;63(12):1407-13. Biochemistry (Mosc). 1998. PMID: 9916158
-
Quinone-dependent alcohol dehydrogenases and FAD-dependent alcohol oxidases.Biochemistry (Mosc). 2012 Aug;77(8):843-56. doi: 10.1134/S0006297912080056. Biochemistry (Mosc). 2012. PMID: 22860906 Review.
-
Biosynthesis and assembly of alcohol oxidase, a peroxisomal matrix protein in methylotrophic yeasts: a review.Yeast. 1991 Apr;7(3):195-209. doi: 10.1002/yea.320070302. Yeast. 1991. PMID: 1882546 Review.
Cited by
-
Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.Acta Crystallogr D Struct Biol. 2021 Jun 1;77(Pt 6):755-775. doi: 10.1107/S2059798321003533. Epub 2021 May 14. Acta Crystallogr D Struct Biol. 2021. PMID: 34076590 Free PMC article.
-
Implications of evolutionary engineering for growth and recombinant protein production in methanol-based growth media in the yeast Pichia pastoris.Microb Cell Fact. 2017 Mar 17;16(1):49. doi: 10.1186/s12934-017-0661-5. Microb Cell Fact. 2017. PMID: 28302114 Free PMC article.
-
Functional Classification of Super-Large Families of Enzymes Based on Substrate Binding Pocket Residues for Biocatalysis and Enzyme Engineering Applications.Front Bioeng Biotechnol. 2021 Aug 2;9:701120. doi: 10.3389/fbioe.2021.701120. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34409021 Free PMC article.
-
The GMC superfamily of oxidoreductases revisited: analysis and evolution of fungal GMC oxidoreductases.Biotechnol Biofuels. 2019 May 10;12:118. doi: 10.1186/s13068-019-1457-0. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31168323 Free PMC article.
-
Structure-Based Engineering of Phanerochaete chrysosporium Alcohol Oxidase for Enhanced Oxidative Power toward Glycerol.Biochemistry. 2018 Oct 30;57(43):6209-6218. doi: 10.1021/acs.biochem.8b00918. Epub 2018 Oct 16. Biochemistry. 2018. PMID: 30272958 Free PMC article.
References
-
- Kiess M, Hecht HJ, Kalisz HM. Glucose oxidase from Penicillium amagasakiense. Primary structure and comparison with other glucose-methanol-choline (GMC) oxidoreductases. Eur J Biochem. 1998;252(1):90–9. . - PubMed
-
- Roggenkamp R, Janowicz Z, Stanikowski B, Hollenberg CP. Biosynthesis and regulation of the peroxisomal methanol oxidase from the methylotrophic yeast Hansenula polymorpha. Mol Gen Genet. 1984;194(3):489–93. . - PubMed
-
- Sibirny AA, Titorenko VI, Efremov BD, Tolstorukov II. Multiplicity of Mechanisms of Carbon Catabolite Repression Involved in the Synthesis of Alcohol Oxidase in the Methylotrophic Yeast Pichia-Pinus. Yeast. 1987;3(4):233–41. 10.1002/yea.320030404 . - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

