Unique Loss of the PYHIN Gene Family in Bats Amongst Mammals: Implications for Inflammasome Sensing
- PMID: 26906452
- PMCID: PMC4764838
- DOI: 10.1038/srep21722
Unique Loss of the PYHIN Gene Family in Bats Amongst Mammals: Implications for Inflammasome Sensing
Abstract
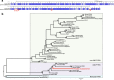
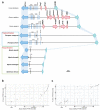
Recent genomic analysis of two bat species (Pteropus alecto and Myotis davidii) revealed the absence of the PYHIN gene family. This family is recognized as important immune sensors of intracellular self and foreign DNA and activators of the inflammasome and/or interferon pathways. Further assessment of a wider range of bat genomes was necessary to determine if this is a universal pattern for this large mammalian group. Here we expanded genomic analysis of this gene family to include ten bat species. We confirmed the complete loss of this gene family, with only a truncated AIM2 remaining in one species (Pteronotus parnellii). Divergence of the PYHIN gene loci between the bat lineages infers different loss-of-function histories during bat evolution. While all other major groups of placental mammals have at least one gene member, only bats have lost the entire family. This removal of inflammasome DNA sensors may indicate an important adaptation that is flight-induced and related, at least in part, to pathogen-host co-existence.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

