Metabolic prediction of important agronomic traits in hybrid rice (Oryza sativa L.)
- PMID: 26907211
- PMCID: PMC4764848
- DOI: 10.1038/srep21732
Metabolic prediction of important agronomic traits in hybrid rice (Oryza sativa L.)
Abstract
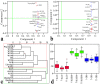
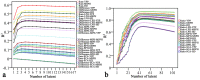
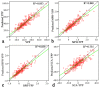
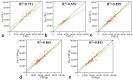
Hybrid crops have contributed greatly to improvements in global food and fodder production over the past several decades. Nevertheless, the growing population and changing climate have produced food crises and energy shortages. Breeding new elite hybrid varieties is currently an urgent task, but present breeding procedures are time-consuming and labour-intensive. In this study, parental metabolic information was utilized to predict three polygenic traits in hybrid rice. A complete diallel cross population consisting of eighteen rice inbred lines was constructed, and the hybrids' plant height, heading date and grain yield per plant were predicted using 525 metabolites. Metabolic prediction models were built using the partial least square regression method, with predictive abilities ranging from 0.858 to 0.977 for the hybrid phenotypes, relative heterosis, and specific combining ability. Only slight changes in predictive ability were observed between hybrid populations, and nearly no changes were detected between reciprocal hybrids. The outcomes of prediction of the three highly polygenic traits demonstrated that metabolic prediction was an accurate (high predictive abilities) and efficient (unaffected by population genetic structures) strategy for screening promising superior hybrid rice. Exploitation of this pre-hybridization strategy may contribute to rice production improvement and accelerate breeding programs.
Figures
References
-
- Yu C. Y., Hu S. W., Zhao H. X., Guo A. G. & Sun G. L. Genetic distances revealed by morphological characters, isozymes, proteins and RAPD markers and their relationships with hybrid performance in oilseed rape (Brassica napus L.). Theor Appl Genet 110, 511–518 (2005). - PubMed
-
- Reif J. C., Zhao Y., Würschum T., Gowda M., Hahn V. & Léon J. Genomic prediction of sunflower hybrid performance. Plant Breed 132, 107–114 (2013).
-
- Schrag T. A. et al. Molecular marker-based prediction of hybrid performance in maize using unbalanced data from multiple experiments with factorial crosses. Theor Appl Genet 118, 741–751 (2009). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

