Identification of Known and Novel Recurrent Viral Sequences in Data from Multiple Patients and Multiple Cancers
- PMID: 26907326
- PMCID: PMC4776208
- DOI: 10.3390/v8020053
Identification of Known and Novel Recurrent Viral Sequences in Data from Multiple Patients and Multiple Cancers
Abstract
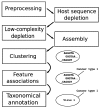
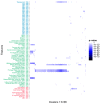
Virus discovery from high throughput sequencing data often follows a bottom-up approach where taxonomic annotation takes place prior to association to disease. Albeit effective in some cases, the approach fails to detect novel pathogens and remote variants not present in reference databases. We have developed a species independent pipeline that utilises sequence clustering for the identification of nucleotide sequences that co-occur across multiple sequencing data instances. We applied the workflow to 686 sequencing libraries from 252 cancer samples of different cancer and tissue types, 32 non-template controls, and 24 test samples. Recurrent sequences were statistically associated to biological, methodological or technical features with the aim to identify novel pathogens or plausible contaminants that may associate to a particular kit or method. We provide examples of identified inhabitants of the healthy tissue flora as well as experimental contaminants. Unmapped sequences that co-occur with high statistical significance potentially represent the unknown sequence space where novel pathogens can be identified.
Keywords: assay contamination; cancer causing viruses; next generation sequencing; novel sequence identification; oncoviruses; sequence clustering; taxonomic characterisation.
Figures


Similar articles
-
Cataloguing the taxonomic origins of sequences from a heterogeneous sample using phylogenomics: applications in adventitious agent detection.PDA J Pharm Sci Technol. 2014 Nov-Dec;68(6):602-18. doi: 10.5731/pdajpst.2014.01023. PDA J Pharm Sci Technol. 2014. PMID: 25475635
-
Detecting viral sequences in NGS data.Curr Opin Virol. 2019 Dec;39:41-48. doi: 10.1016/j.coviro.2019.07.010. Epub 2019 Aug 26. Curr Opin Virol. 2019. PMID: 31465960 Review.
-
Sensitive detection of viral transcripts in human tumor transcriptomes.PLoS Comput Biol. 2013;9(10):e1003228. doi: 10.1371/journal.pcbi.1003228. Epub 2013 Oct 3. PLoS Comput Biol. 2013. PMID: 24098097 Free PMC article.
-
Viral sequences in human cancer.Virology. 2018 Jan 1;513:208-216. doi: 10.1016/j.virol.2017.10.017. Epub 2017 Nov 5. Virology. 2018. PMID: 29107929 Free PMC article.
-
Advances in the application of high-throughput sequencing in invertebrate virology.J Invertebr Pathol. 2017 Jul;147:145-156. doi: 10.1016/j.jip.2017.02.006. Epub 2017 Feb 27. J Invertebr Pathol. 2017. PMID: 28249815 Review.
Cited by
-
High diversity of picornaviruses in rats from different continents revealed by deep sequencing.Emerg Microbes Infect. 2016 Aug 17;5(8):e90. doi: 10.1038/emi.2016.90. Emerg Microbes Infect. 2016. PMID: 27530749 Free PMC article.
-
Metagenomic Identification of Viral Sequences in Laboratory Reagents.Viruses. 2021 Oct 21;13(11):2122. doi: 10.3390/v13112122. Viruses. 2021. PMID: 34834931 Free PMC article.
-
Tumour virology in the era of high-throughput genomics.Philos Trans R Soc Lond B Biol Sci. 2017 Oct 19;372(1732):20160265. doi: 10.1098/rstb.2016.0265. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28893932 Free PMC article. Review.
-
A virome-wide clonal integration analysis platform for discovering cancer viral etiology.Genome Res. 2019 May;29(5):819-830. doi: 10.1101/gr.242529.118. Epub 2019 Mar 14. Genome Res. 2019. PMID: 30872350 Free PMC article.
-
The Human Virome: Implications for Clinical Practice in Transplantation Medicine.J Clin Microbiol. 2017 Oct;55(10):2884-2893. doi: 10.1128/JCM.00489-17. Epub 2017 Jul 19. J Clin Microbiol. 2017. PMID: 28724557 Free PMC article. Review.
References
-
- Woo P.C.Y., Lau S.K.P., Chu C., Chan K., Tsoi H., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W., et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

